 01Research Overview
01Research Overview
With the rapid development of molecular diagnostic technologies, digital CRISPR technology has become an important tool for nucleic acid detection due to its single-molecule counting and absolute quantification capabilities. However, traditional digital CRISPR platforms face significant challenges in several aspects. Firstly, most existing platforms adopt equal-volume microcavity designs, which typically cover a dynamic range of only about 3 orders of magnitude, making it difficult to accurately detect both high and low concentration samples simultaneously. Secondly, traditional multiplex target detection platforms require multiple fluorescent channels or multiple Cas nucleases to distinguish targets, leading to increased system complexity and higher costs. To address these issues, Professor Mu Ying’s team at Zhejiang University developed a microfluidic chip called WDRM-dCRISPR. By introducing a multi-volume microcavity array and six independent detection zones, this platform successfully overcomes the bottlenecks of traditional chips, achieving a dynamic range from 9×10¹ to 1.8×10⁶ copies·mL⁻¹, while supporting multiplex target detection. This technological breakthrough provides new ideas for widespread portable nucleic acid detection. The related results were published under the title “Wide Dynamic Range Multiplex Digital Clustered Regularly Interspaced Short Palindromic Repeat Chip for Rapid Detection and Absolute Quantification of Nucleic Acids” in ACS Nano (DOI: 10.1021/acsnano.5c03382).
02
Research Content
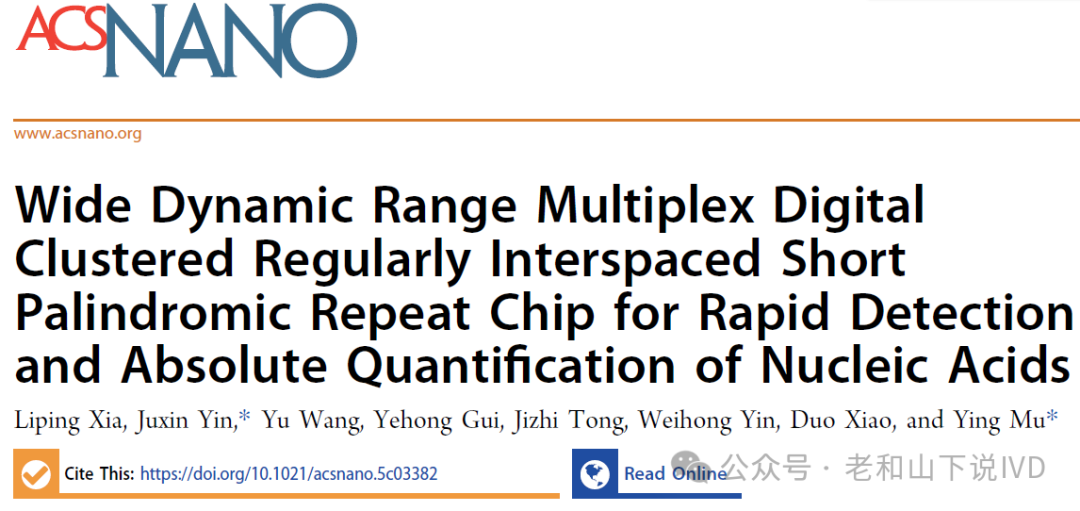
The detection platform first releases the nucleic acids of the target pathogens through thermal lysis and loads them into a chip pre-filled with complete reaction components. The chip is then placed in an automated temperature-controlled fluorescence imaging device for analysis. When the target nucleic acid is present, the Cas12a crRNA complex specifically recognizes the target nucleic acid fragment and activates its trans-cleavage activity, randomly cleaving the ssDNA report molecules modified with fluorescent and quenching groups, generating corresponding fluorescence signals and forming positive reaction cavities. Conversely, if the target nucleic acid fragment is absent, the trans-cleavage activity of Cas12a is not activated, and no fluorescence signal is produced. Ultimately, by counting the number of positive microcavities in each volume reaction zone and combining it with the most probable number (MPN) theory, the absolute concentration of nucleic acids is calculated, achieving high sensitivity and precision in nucleic acid quantification.
The design innovation of the WDRM-dCRISPR chip lies in its multi-volume microcavity array and multi-detection zone construction. Each detection zone (DZ) contains four different volumes of microcavities, with volume ratios of 1 : 8 : 32 : 64, totaling 240 microcavity units. This design allows the chip to effectively cover a wide sample range from low to high concentrations, enhancing the platform’s dynamic range and sensitivity. Through this multi-volume design, low concentration samples can be captured through larger volume microcavities, thereby improving sensitivity, while high concentration samples effectively avoid signal saturation through smaller volume microcavities, ensuring accurate detection of high concentration samples. As shown in Figure 1, the overall layout design of the chip is displayed. Each detection zone is arranged into six independent areas, each containing microcavities of different volumes to ensure effective reactions when processing samples of varying concentrations. Each microcavity is connected to the main inlet, allowing samples to be evenly distributed to each area through negative pressure. The design layout in the figure reflects the diversity of microcavities within each detection zone and how different volumes are used to handle samples from low to high concentrations, expanding the dynamic range of the chip.
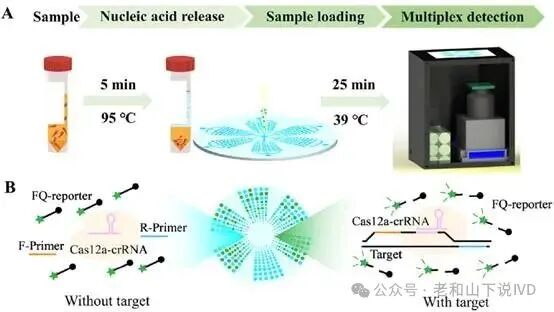
Figure 1. Schematic of the detection principle of the WDRM-dCRISPR chip
The microcavity array of the chip is made through a three-layer composite material structure. The bottom layer is an optically transparent glass substrate, providing mechanical support and facilitating optical imaging; the middle layer is a microstructure layer, creating microchannels and microcavities through soft lithography; the top layer is a support layer, sealing the reaction cavities and ensuring the stability and closure of the microcavities. As shown in Figure 2, the figure details the microcavity array and structural hierarchy of the chip. Under this structural design, the size and shape of each microcavity can be precisely controlled, ensuring the stability of the reaction process and reducing potential errors in experiments. To further enhance the operational convenience of the chip and reduce operational complexity, the researchers designed a secondary inlet for pre-injecting reagents. Each detection zone’s secondary inlet can pre-inject reaction components, ensuring that the reaction components in each area are correctly configured before sample loading. The main inlet is used for sample loading, and samples are automatically distributed to each detection zone through negative pressure. The collaborative mechanism between the secondary and main inlets ensures automatic sample distribution and independent reagent loading. Through this design, the stability of the reaction zones is guaranteed, avoiding the risk of cross-contamination.
Figure 2. Design diagram of the WDRM-dCRISPR chip
The WDRM-dCRISPR chip demonstrated extremely high sensitivity and specificity in performance evaluation. As shown in Figure 3, the researchers used marker genes from five pathogens for detection, namely Salmonella (invA), E. coli (rfbE), L. monocytogenes (hlyA), V. parahemolyticus (tlh), and S. flexi (ipaH). The experimental results indicated that the chip could accurately detect all targets within the range of 9×10¹ to 1×10⁵ copies·mL⁻¹, with no cross-interference among the five targets. As shown in Figure 4, the figure displays the detection results of different concentrations of targets on the chip. Through quantitative experiments in samples of varying concentrations, the quantitative results of the chip were highly consistent with traditional qPCR methods, and it could accurately detect low concentration samples, demonstrating its superior sensitivity. Particularly in low concentration samples, the WDRM-dCRISPR chip successfully detected pathogens that were missed by qPCR, further proving the chip’s advantages in complex samples.
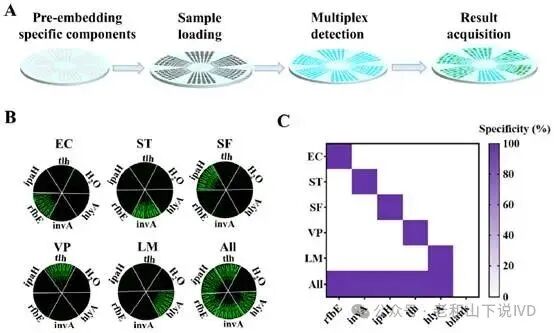
Figure 3. Specificity characterization of the WDRM-dCRISPR chip
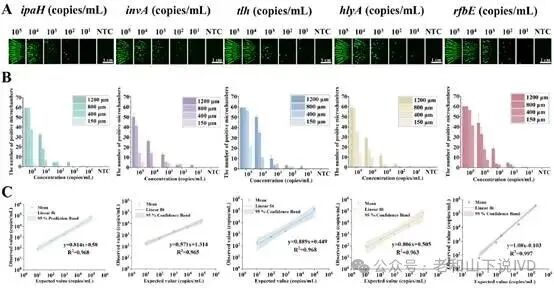
Figure 4. Sensitivity and dynamic range of the WDRM-dCRISPR chip
In actual sample validation, the researchers selected complex matrices such as milk for detection. The experimental results indicated that the WDRM-dCRISPR chip could successfully detect low concentration pathogens in complex samples and exhibited superior inhibition resistance and stability compared to qPCR. As shown in Figure 5, the figure compares the detection results of the chip in milk samples with the qPCR method. At low concentrations, the chip successfully detected pathogens, demonstrating stronger sensitivity and adaptability. Ultimately, the detection time of the WDRM-dCRISPR chip was optimized, requiring only 30 min from sample loading to result output, significantly improving detection efficiency. Through a portable temperature-controlled imaging system, the chip can complete the entire process from sample loading to final result output in a short time, showing broad potential for field applications.
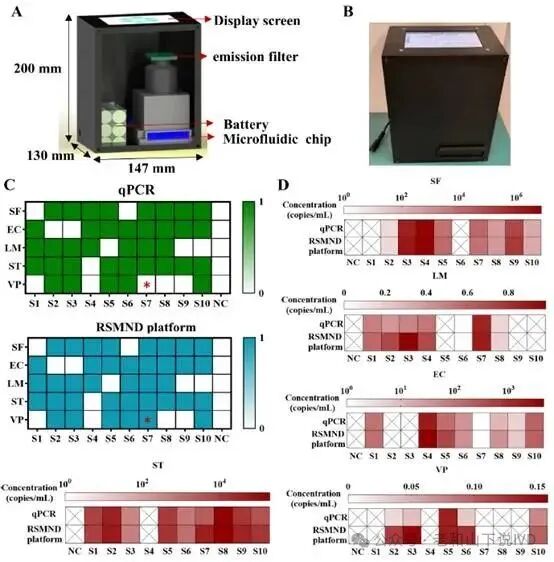
Figure 5. Evaluation of actual detection performance03Research Summary
The WDRM-dCRISPR chip successfully overcomes the limitations of traditional CRISPR detection platforms in dynamic range and multiplex target detection through its innovative multi-volume microcavity array design. In actual sample detection, the platform demonstrated high sensitivity, strong specificity, and excellent interference resistance, capable of completing the entire process from sample loading to result output within 30 min, with broad potential for field applications. The design and optimization of this platform provide new ideas and paradigms for future multiplex, one-pot CRISPR detection platforms.
Note: This article is a report on the latest scientific research developments. If there are any conflicts with the original authors, please contact the author of this public account for deletion.
 THE END
THE END Overlooking the Mountains
Overlooking the Mountains Author|Harold Proofreader|HD Push|HDReferences>>
Author|Harold Proofreader|HD Push|HDReferences>>
1. Wide Dynamic Range Multiplex Digital Clustered Regularly Interspaced Short Palindromic Repeat Chip for Rapid Detection and Absolute Quantification of Nucleic Acids. ACS Nano, 2025, DOI: 10.1021/acsnano.5c03382.
