
◆ ◆ ◆ ◆
Sci Adv | Development of uCoTargetX Technology for Single-Cell Multi-Protein Modification and Transcription Detection
Technological innovation is a crucial engine for modern life sciences research. Each iteration and update of biotechnology brings disruptive changes to the life sciences field. In recent years, single-cell sequencing technology has developed rapidly, propelling scientists worldwide into a fast track to uncover the mysteries of life. Single-cell multi-omics technology is an important tool for studying cell-specific chromatin states and gene expression regulatory mechanisms. High-precision analysis of multi-dimensional epigenetic landscapes at the single-cell level has become a focal point in the field. To investigate the epigenetic regulatory mechanisms of cell fate in complex life systems, single-cell multi-dimensional histone modification detection technology has begun to emerge in the past two years【1-5】, but existing technologies still have significant limitations: (1) Experimental steps are complex and rely on special reagents and instruments, making it difficult to promote their use in general biomedical laboratories; (2) the number of histone modifications that can be captured simultaneously is limited; (3) the quality of single-cell data is relatively low and lacks true single-cell multi-dimensional epigenomic and transcriptomic co-analysis. Therefore, there is an urgent need to develop a new type of simple-to-use, ultra-high-throughput, high-quality, and highly sensitive single-cell multi-dimensional histone modification detection technology.
On January 3, 2024, the team of He Aibin from the School of Future Technology at Peking University and the Peking University-Tsinghua University Joint Center for Life Sciences published an article titled “Single-cell joint profiling of multiple epigenetic proteins and gene transcription” in the journal Science Advances, reporting a new single-cell multi-dimensional epigenetic detection technology named uCoTarget (ultra-high throughput Combined TAgmenting enRichment for multiple epiGEneTic proteins in the same cells), which can simultaneously capture up to five different protein-chromatin interaction information in a single cell. Based on this, the researchers further optimized and innovated to develop the uCoTargetX technology, which is the first in the world to achieve parallel detection of multiple histone modifications and transcriptomes at the single-cell level, providing a powerful technical tool for in-depth research on the epigenetic regulation of cell fate under various physiological and pathological conditions.
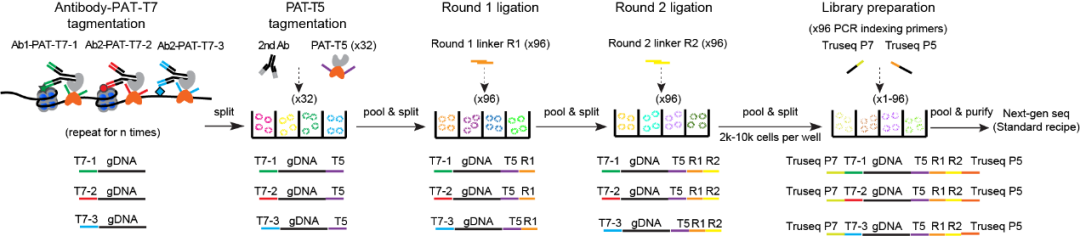
Figure 1: Workflow of the ultra-high throughput single-cell multi-omics technology uCoTarget
Wang Qianhao, a PhD graduate from the School of Future Technology at Peking University, and Researcher Xiong Haiqing from the Institute of Hematology, Chinese Academy of Medical Sciences, are the co-first authors of the paper. Professor He Aibin and Researcher Xiong Haiqing from the Peking University-Tsinghua University Joint Center for Life Sciences are the corresponding authors of this paper. This research was supported by the Stem Cell Special Project of the Ministry of Science and Technology, the National Natural Science Foundation, and the Joint Center for Life Sciences.
Full text link:
https://www.science.org/doi/10.1126/sciadv.adi3664
Appendix: Comparison of characteristics of different single-cell multi-protein modification technologies

References:
1. Gopalan S, Wang Y, Harper N W, et al. Simultaneous profiling of multiple chromatin proteins in the same cells. Molecular cell, 2021, 81(22): 4736-4746. e5.
2. Stuart T, Hao S, Zhang B, et al. Nanobody-tethered transposition enables multifactorial chromatin profiling at single-cell resolution. Nature Biotechnology, 2023, 41(6): 806-812.
3. Bartosovic M, Castelo-Branco G. Multimodal chromatin profiling using nanobody-based single-cell CUT&Tag. Nature Biotechnology, 2023, 41(6): 794-805.
4. Meers M P, Llagas G, Janssens D H, et al. Multifactorial profiling of epigenetic landscapes at single-cell resolution using MulTI-Tag. Nature Biotechnology, 2023, 41(5): 708-716.
5. Lochs S J A, van der Weide R H, de Luca K L, et al. Combinatorial single-cell profiling of major chromatin types with MAbID. Nature Methods, 2023: 1-11.
6. Wang Q, Xiong H, Ai S, et al. CoBATCH for high-throughput single-cell epigenomic profiling. Molecular cell, 2019, 76(1): 206-216. e7.
7. Ai S, Xiong H, Li C C, et al. Profiling chromatin states using single-cell itChIP-seq. Nature Cell Biology, 2019, 21(9): 1164-1172.
8. Xiong H, Luo Y, Wang Q, et al. Single-cell joint detection of chromatin occupancy and transcriptome enables higher-dimensional epigenomic reconstructions. Nature Methods, 2021, 18(6): 652-660.

He Aibin
Professor at the School of Future Technology, Peking University
PI at the Peking University-Tsinghua University Joint Center for Life Sciences
The He Aibin research group primarily studies the mechanisms of cell lineage origin and fate regulation during embryonic development and disease processes. Our research focuses on the following three areas: 1. The epigenetic regulatory mechanisms of organ stem cell fate differentiation from the three germ layers during embryonic development. We develop single-cell multi-omics sequencing technologies (transcriptome and multi-dimensional epigenome) to explore key silencer element sets, enhancer sets, and their regulatory transcription factor networks that determine cell fate. 2. The application of novel single-cell epigenomics technologies in early disease diagnosis. We focus on blood and heart diseases. 3. In vivo real-time imaging and digital cell lineage tracing of the cellular mechanisms of embryonic morphogenesis. Using mouse embryos as models, we utilize self-built light-sheet microscopy and image processing techniques, combined with cell lineage tracing, to track stem cell behavior (proliferation, differentiation, migration, and distribution) and map digital cell lineage landscapes.
This article is reproduced from the WeChat public account “Joint Center for Life Sciences”
