Click the “Nanjing University IFB” above to follow us!
Hello everyone, today I would like to share a recent article published in the Journal of the American Chemical Society:Revision of the Formycin A and Pyrazofurin Biosynthetic Pathways Reveals Specificity for D‑Glutamic Acid and a Cryptic N‑Acylation Step During Pyrazole Core Formation, with the corresponding author being Professor Hung-wen Liu from the University of Texas at Austin.Professor Hung-wen Liu has long been engaged in fundamental research at the intersection of chemistry and biology, focusing on the reaction mechanisms of novel enzymes in the biosynthetic pathways of important natural products and designing methods to control or modify enzyme functions. This article revises the biosynthetic pathways of Formycin A and Pyrazofurin, discovering that the key enzymes for the formation of their pyrazole ring, PyfG and ForJ, specifically recognize D-glutamic acid instead of the previously thought L-glutamic acid, and revealing a cryptic N-acylation step catalyzed by an ATP-dependent ligase, which provides a necessary protective mechanism for subsequent oxidation and cyclization reactions; these findings not only correct previous errors but also provide new insights into the synthesis mechanisms of rare pyrazole rings in natural products.
Both Formycin A and Pyrazofurin are linked to the pyrazole ring through a C-glycosidic bond and belong to the class of pyrazole-C-nucleoside antibiotic compounds. The chemical structures of Formycin A and Pyrazofurin not only feature characteristic C-glycosidic linkages but also possess an unusual pyrazole-derived nucleobase. The construction of the C-glycosidic bond is catalyzed by the C-glycosyltransferases ForT and PyfQ (Figure 1). However, the synthesis of the shared pyrazole core in Formycin A and Pyrazofurin remains largely unknown.
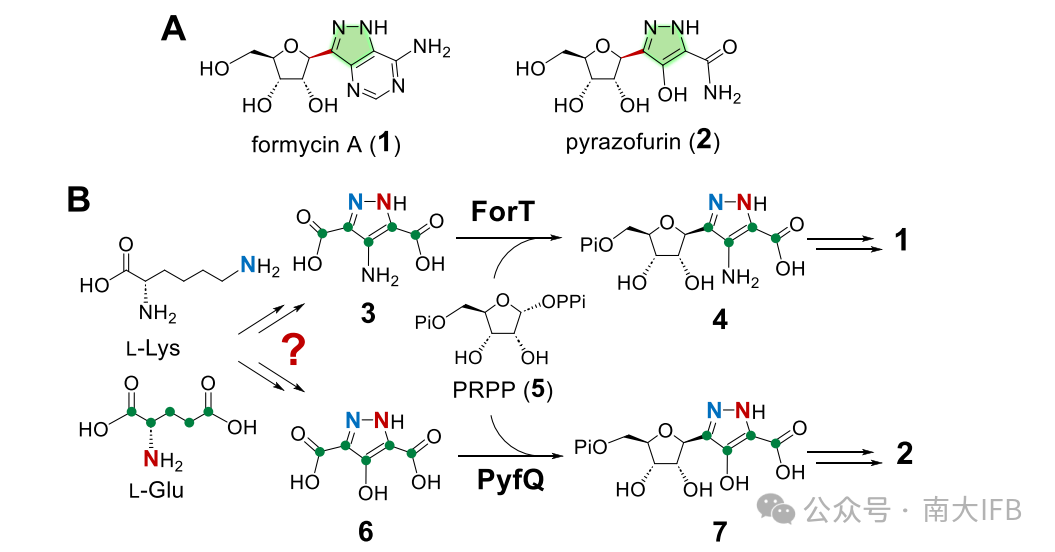
Figure 1: Structures of Formycin A and Pyrazofurin, with the pyrazole ring highlighted in green and the C-glycosidic bond marked in red. (B) C-glycosylation reaction scheme catalyzed by ForT and PyfQ.
Previous studies suggested that ForJ/PyfG (homologous to Spb40) catalyze the N⁶-hydroxylation of L-lysine (9) and its coupling with L-Glu (producing L-13). However, this article demonstrates through in vitro enzyme reactions that ForJ/PyfG specifically recognizes D-Glu, producing the intermediate D-13. When using D-Glu, ForJ/PyfG catalyzes the formation of D-13 (m/z 292.1499 [M+H]+), confirmed by Fmoc derivatization and NMR.L-Glu cannot produce the product (Figure 2), and the activity detected in earlier studies may have stemmed from impurities of D-Glu (for instance, the presence of 0.5% D-Glu in L-Glu could yield detectable amounts of product).
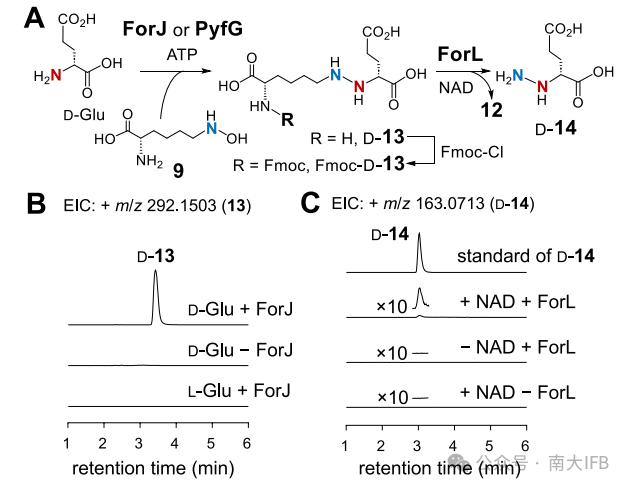
Figure 2: (A) Reactions catalyzed by ForJ/PyfG and ForL. (B) LCMS analysis of the coupling reaction of ForJ and (C) ForJ-ForL.
In vitro enzyme reaction experiments confirmed that ForM acts as an ATP-grasp ligase, catalyzing the N2′ acylation of α-hydrazinyl D-glutamic acid (D-14) (Figure 3).ForM accepts five amino acids such as glycine, L-serine, and L-threonine as acyl donors, producing D-14 analogs (e.g., D-41, m/z 220.0929 [M+H]+). The FAD-dependent oxidoreductase ForR specifically recognizes D- acylation products (e.g., D-41), catalyzing the dehydrogenation of the Cα-N bond to generate the hydrazone intermediate 42 (m/z 218.0768 [M+H]+)(Figure 3).L- acylation products (L-41) cannot be recognized by ForR, explaining the failure of earlier experiments.
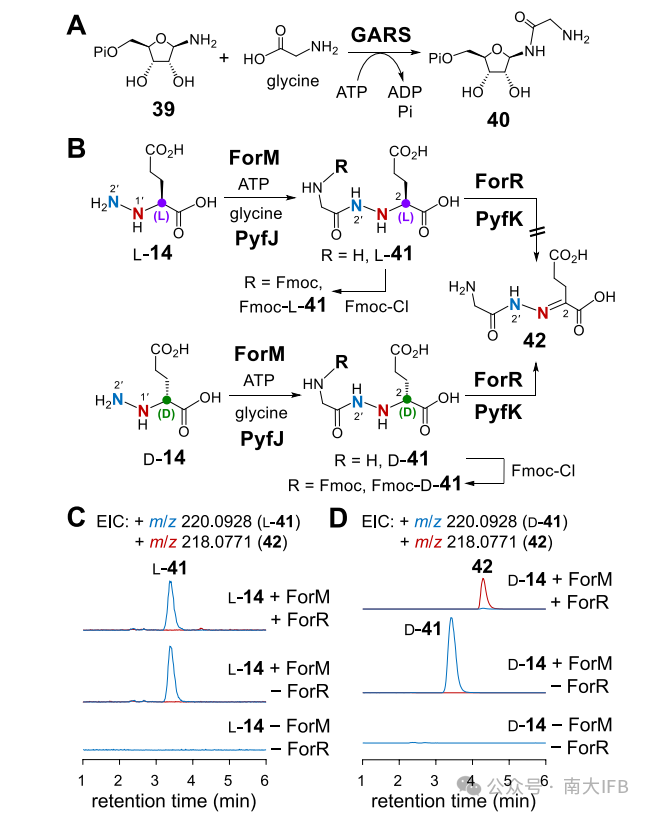
Figure 3: (A) Reaction catalyzed by guanylyltransferase (GARS).(B) Reactions catalyzed by ForM/PyfJ and ForR/PyfK using L-14 or D-14 as substrates. (C) LCMS analysis of the coupling reaction of ForM and ForM-ForR.
The authors validated the oxidative release function of ForL/PyfI through in vitro reactions (requiring NAD cofactor): NAD-dependent oxidoreductase ForL catalyzes the conversion of D-13 to D-14, releasing L-2-aminohexanedioic acid-6-aldehyde (12) (Figure 2).The subsequent oxidation and cyclization mechanisms (Figure 4): C3/C4 hydroxylation: FMN-dependent monooxygenases (ForD/E or PyfM/N) catalyze the C3 hydroxylation of the hydrazone intermediate 42, generating 45 (Formycin branch) or directly generating 49 (Pyrazofurin branch). Amination modification: The unique amino group of Formycin is introduced by the PLP-dependent aminotransferase ForI. Dehydrogenation and hydrolysis: Rieske oxidoreductase ForU/PyfB and dehydrogenase ForS/PyfO catalyze C4 hydroxylation and dehydrogenation, ultimately hydrolyzing the amide bond by ForQ/PyfA, triggering the cyclization of the pyrazole ring (52→3 or 54→6) (Figure 4).
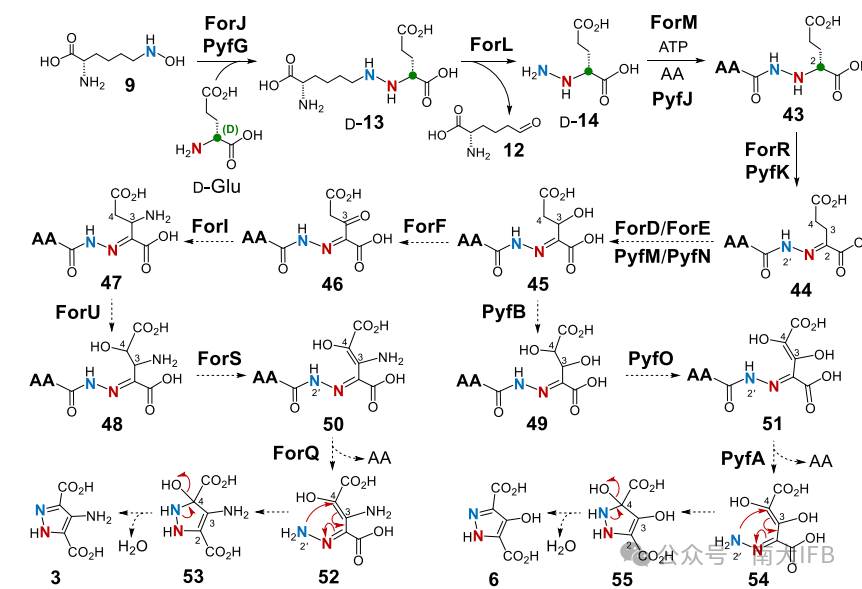
Figure 4: Proposed pathways for the formation of the pyrazole ring in the biosynthesis of Formycin A and Pyrazofurin.
In summary: This article revises the biosynthetic pathways of the natural products Formycin A and Pyrazofurin, revealing the key mechanisms for pyrazole ring formation. The study found that the enzymes responsible for N-N bond formation, ForJ and PyfG, specifically utilize D-glutamic acid (rather than the previously thought L-glutamic acid) as a substrate, producing the intermediate D-13; subsequently, the NAD-dependent oxidoreductase ForL catalyzes the release of α-hydrazinyl D-glutamic acid (D-14). Through the ATP-grasp ligase ForM/PyfJ, N-acylation (such as glycine transfer) of D-14 occurs, forming the intermediate D-41, which is then specifically catalyzed by the FAD-dependent oxidoreductase ForR/PyfK to dehydrogenate and generate the hydrazone intermediate. The study further proposes the enzymatic mechanisms for subsequent oxidation, hydroxylation, and cyclization steps, clarifying the specific role of D-glutamic acid and the cryptic N-acylation step’s critical contribution to the assembly of the pyrazole ring, providing new perspectives for understanding the biosynthesis of pyrazole-containing natural products.
Article Title: Revision of the Formycin A and Pyrazofurin Biosynthetic Pathways Reveals Specificity for D‑Glutamic Acid and a Cryptic N‑Acylation Step During Pyrazole Core Formation
Full text link: https://doi.org/10.1021/jacs.5c01277
Compiled by: Wang Zirui
Long press the image below to recognize the QR code in the image and easily follow us!
