Currently, an increasing number of papers involve DFT calculations, with the Molecular Electrostatic Potential (MEP) being used more widely. So how can we obtain high-quality MEP maps? Let’s follow the steps outlined below.
1
Software Installation
VMD Installation:
Multiwfn Installation:
2
Software Preparation
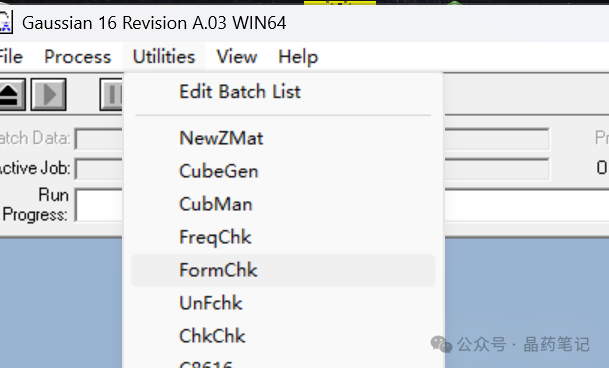
Locate the chk file generated after Gaussian optimization. Open Gaussian, click on Utilities, then FormChk, and input the chk file. This will generate an fck file in the original folder. Rename this file to 1.fck and place it in the folder where Multiwfn is located.
First, find the locations of the VMD and Multiwfn folders. If you cannot find them, right-click the VMD icon and select ‘Open file location’.
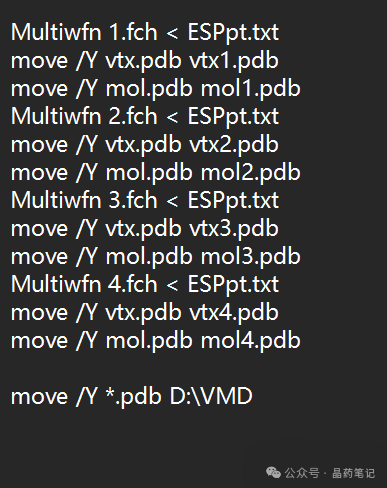
Configure the Multiwfn files: In the Multiwfn examples\drawESP directory, copy the files ESPiso.bat, ESPpt.bat, ESPext.bat, ESPiso.txt, ESPpt.txt, and ESPext.txt, and paste them into the examples folder (note that it should be the examples folder, not inside examples; incorrect placement may cause crashes). Use a text editor to edit ESPiso.bat, ESPpt.bat, and ESPext.bat, changing the first line to 1.fck and the last line to the path of the VMD folder, as shown in the image:
Configure VMD: In the Multiwfn examples\drawESP directory, copy ESPiso.vmd, ESPiso2.vmd, ESPpt.vmd, ESPpt2.vmd, and ESPext.vmd to the VMD folder.
Open the vmd.rc file in that directory using a text editor.

Paste the following lines at the end of the text:
proc iso {} {source ESPiso.vmd}proc iso2 {} {source ESPiso2.vmd}proc pt {} {source ESPpt.vmd}proc pt2 {} {source ESPpt2.vmd}proc ext {} {source ESPext.vmd}
After completing these steps, we have configured all the software.
3
Generating Molecular Electrostatic Potential Maps
Press Ctrl and select ESPiso.bat, ESPpt.bat, and ESPext.bat simultaneously, then right-click to open them. The three modules will start running.

Once the process is complete, the window will close automatically, and you will find three files in the VMD directory indicating that the software has finished running.
Next, double-click to open the vmd.exe software.
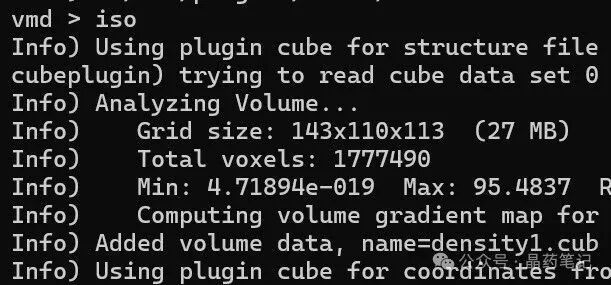
In the VMD text window, type ‘iso’ to display the electronic density isosurface, showing the molecular van der Waals surface electrostatic potential distribution.
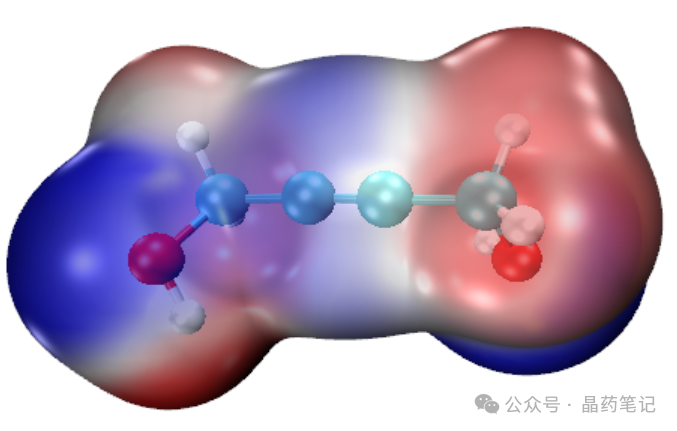
Continue by typing ‘ext’ to obtain the extreme points.
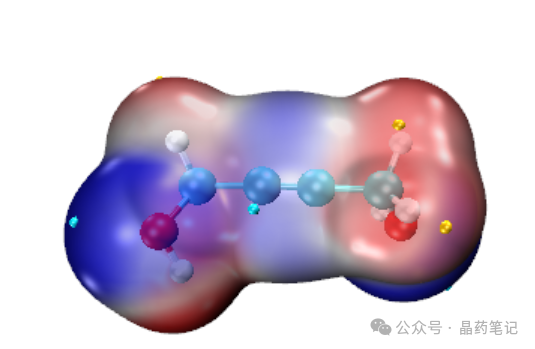
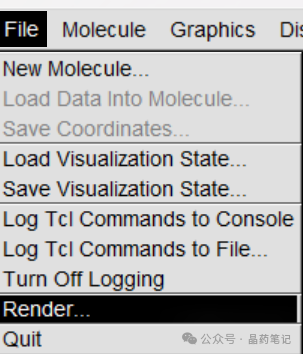
Then, in the VMD window, click File – Render to save the image. It is recommended to select Tachyon for rendering. Save the image in the VMD directory.
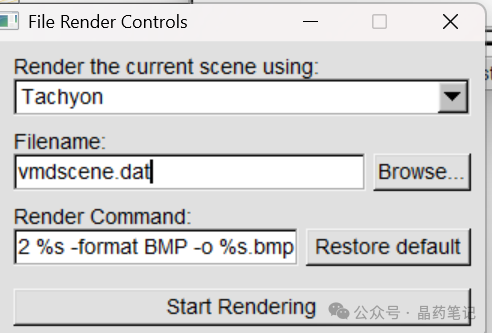
Rendering: Finally, open VMDrender_full in the VMD directory to render the image. You will obtain the rendered molecular electrostatic potential image full.bmp in the VMD directory. You can modify this image in Photoshop by adding extreme points to use it as a figure in your paper!

Disclaimer: This content is a summary of the Multiwfn software manual published at the entrance of the Thinker Community: Quantum Chemistry · Molecular Simulation · 2D, along with examples for demonstration.
This operation is relatively simple and does not provide a detailed explanation of the principles. If you are interested, please visit the Thinker Community: Quantum Chemistry · Molecular Simulation · 2D website for a detailed and systematic study.
Website: http://sobereva.com/
Additionally, if you use the Multiwfn software in your paper, please cite the developer’s paper. Detailed citation requirements can be found at:
That’s all for today’s sharing. I wish everyone success in publishing their papers!