
Hello everyone, today I would like to share a paper published in 2023 in Talanta, titled “Computer Vision-Based Artificial Intelligence-Mediated Encoding-Decoding for Multiplexed Microfluidic Digital Immunoassay.” This paper develops a multidimensional digital immunoassay through particle-based encoding and AI-based decoding, achieving high sensitivity and convenient multiplex detection. The encoded information in the characteristics of microspheres, including their size, quantity, and color, allows for the simultaneous identification and accurate quantification of multiple targets. AI based on computer vision can analyze microscopic images for information decoding and visually output identification results. Furthermore, optical microscopy imaging can be well integrated with microfluidic platforms, allowing for encoding and decoding through computer vision-based AI. This microfluidic digital immunoassay can simultaneously analyze various inflammatory markers and antibiotics within 30 minutes, with high sensitivity and a detection range from pg/mL to μg/mL, showing great potential as a next-generation intelligent biosensor for multiplexed biological assays. The first author of this paper is Weiqi Zhao, and the corresponding author is Yiping Chen.
Background Introduction
There is a growing demand for ultra-sensitive and absolute quantitative detection in clinical diagnostics, driven by food safety and environmental monitoring, which promotes the development of digital immunoassays. In digital immunoassays, individual or a few reporter gene signals are separated for independent reading, enabling target quantification, improving sensitivity, and achieving absolute quantification. Currently, there are mainly digital immunoassays based on microplates, droplets, and fluorophores, with multiplex digital immunoassays receiving attention. However, assays based on microplates and droplets require microfabrication or microfluidic devices, which are costly and complex, making them unsuitable for immediate detection; assays based on fluorophores do not require these but need fluorescence microscopy and laser excitation. Immunoassays with multiplex detection capabilities are crucial for early diagnosis, but multiplex digital immunoassays are time-consuming and require complex and expensive instruments. Therefore, it is meaningful to develop widely accessible, low-cost, multiplex detection, and high-throughput digital immunoassays.
Artificial intelligence is driving the development of biomedicine and has broad prospects in the field of in vitro diagnostics, outperforming human experts in certain tasks of medical image diagnosis. Computer vision, as a field of AI, performs excellently in tasks such as image classification. Given that optical probes are widely used for reading digital immunoassays, computer vision can enhance their performance. Polystyrene (PS) microspheres are widely used in clinical diagnostics due to their low cost, and AI based on computer vision can identify their size, quantity, and color, providing a feasible route for multiplex and quantitative digital immunoassays.
 Design Principle
Design Principle This paper reports a multidimensional encoding/decoding digital immunoassay using computer vision-based AI for microscopic imaging and particle counting. Microspheres with different characteristics (including color, size, and quantity) can be encoded as signal probes. This paper develops a digital immunoassay for information decoding through microscopic imaging and computer vision-based AI, achieving high sensitivity and convenient multiplex detection (Scheme 1a). PS microspheres can serve as excellent multiplex signal probes, and optical microscopy can identify the encoded information in PS microspheres, including color, size, and quantity. The size and color of PS microspheres can encode two-dimensional information for target identification, while the quantity of PS microspheres can quantify specific targets. Combined with magnetic nanoparticles (MNPs), it can perform magnetic separation of the immunocomplex formed between biotinylated PS microspheres and streptavidin (SA) modified MNPs (Scheme 1b). Additionally, the microfluidic chip is designed as a microreactor for digital immunoassays, specifically integrating mixing reaction channels, magnetic separation channels, image acquisition areas, and waste pools (Scheme 1c). Microfluidic technology, due to its excellent reaction controllability, fast reaction speed, and low reagent consumption, can reduce detection volume and effectively improve sensitivity. Microfluidic devices can be well integrated with optical microscopy imaging that requires only 1-10 μL of sample. Computer vision-based AI has high throughput and parallel processing capabilities, enabling accurate and rapid differentiation of the characteristics of encoded microspheres from microscopic images and quickly counting their quantity and outputting identification results, providing a powerful tool for digital immunoassays.
This paper reports a multidimensional encoding/decoding digital immunoassay using computer vision-based AI for microscopic imaging and particle counting. Microspheres with different characteristics (including color, size, and quantity) can be encoded as signal probes. This paper develops a digital immunoassay for information decoding through microscopic imaging and computer vision-based AI, achieving high sensitivity and convenient multiplex detection (Scheme 1a). PS microspheres can serve as excellent multiplex signal probes, and optical microscopy can identify the encoded information in PS microspheres, including color, size, and quantity. The size and color of PS microspheres can encode two-dimensional information for target identification, while the quantity of PS microspheres can quantify specific targets. Combined with magnetic nanoparticles (MNPs), it can perform magnetic separation of the immunocomplex formed between biotinylated PS microspheres and streptavidin (SA) modified MNPs (Scheme 1b). Additionally, the microfluidic chip is designed as a microreactor for digital immunoassays, specifically integrating mixing reaction channels, magnetic separation channels, image acquisition areas, and waste pools (Scheme 1c). Microfluidic technology, due to its excellent reaction controllability, fast reaction speed, and low reagent consumption, can reduce detection volume and effectively improve sensitivity. Microfluidic devices can be well integrated with optical microscopy imaging that requires only 1-10 μL of sample. Computer vision-based AI has high throughput and parallel processing capabilities, enabling accurate and rapid differentiation of the characteristics of encoded microspheres from microscopic images and quickly counting their quantity and outputting identification results, providing a powerful tool for digital immunoassays.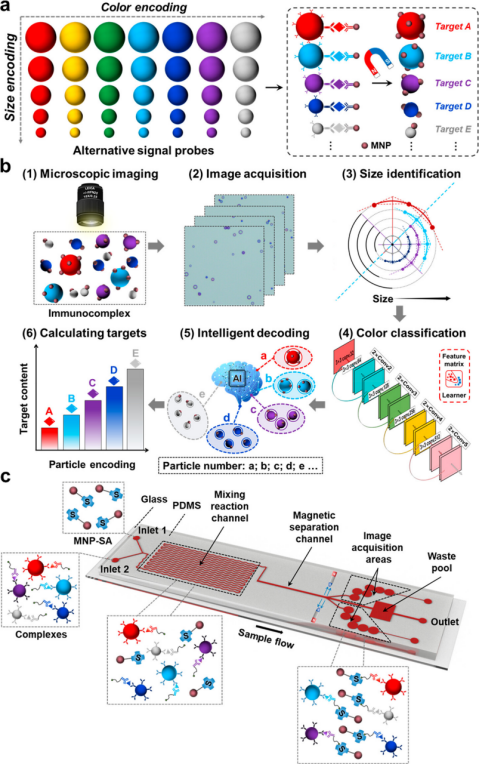 Scheme 1 Schematic diagram of computer vision-based AI for multidimensional encoding/decoding digital immunoassays.Data Introduction
Scheme 1 Schematic diagram of computer vision-based AI for multidimensional encoding/decoding digital immunoassays.Data Introduction
1. Visual Identification of Encoded Microspheres
Eight different sizes and colors of encoded microspheres were selected for visual identification using optical microscopy. The color-encoded microspheres reflect the encoded color light under the light source, and the size and color of PS microspheres can be identified by optical microscopy, allowing for simultaneous encoding of multiple targets for multiplex detection. Different concentrations of size-encoded microspheres were selected, and the quantity of PS microspheres is related to concentration. Combined with the microfluidic chip, optical microscopy is sensitive to the concentration of PS microspheres, capable of recording as low as 1 particle/μL at 200x magnification, achieving high sensitivity imaging and high resolution. The color and size of PS microspheres can encode two-dimensional information for multiplex detection, and the quantity can be quantitatively detected. Compared to xMAP technology, this multifunctional encoding strategy has stronger multiplexing capability, higher sensitivity, and is economical, requiring no expensive complex equipment.
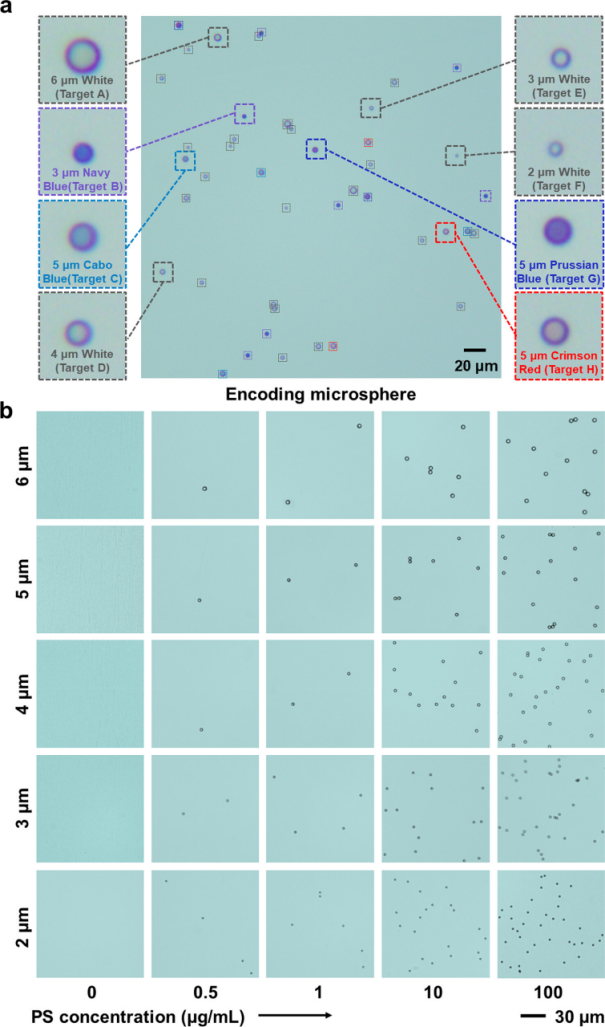
Figure 1 Visual identification of encoded microspheres using optical microscopy.
2. AI Information Decoding Principle Based on Computer Vision
Commercial software and traditional image processing methods cannot perform multidimensional decoding of information, requiring new technology. The AI technology “CAT” based on computer vision was developed to decode multidimensional information using SVM and ResNet algorithms. CAT image processing includes particle size and color decoding, corresponding to the SVM particle size recognition model and ResNet color classification model.
The SVM particle size recognition model first applies bilateral filtering and binarization to the image, using the CHT algorithm to locate and decode the size of microspheres, achieving a classification accuracy of 99.16% for particle size on the test set after optimizing hyperparameters. The ResNet color classification model uses ResNet-18 as the framework to extract color features for classification, and after training, the performance of the color classification model is excellent, with high accuracy and recall rates for the training, validation, and test sets. This strategy effectively classifies encoded microspheres and outputs recognized image decoding.
CAT can reduce the hardware requirements of optical imaging systems. Optical images are affected by aberrations that impact resolution, and it is challenging for the human eye to distinguish multidimensional encoded microsphere features. The high cost of achromatic lenses limits their application. By adding achromatic lenses and ordinary optical lens samples for training, the ResNet color classification model shows good recognition results, but CAT can avoid the impact of aberrations based on deep learning algorithms, broadening the application of image processing.
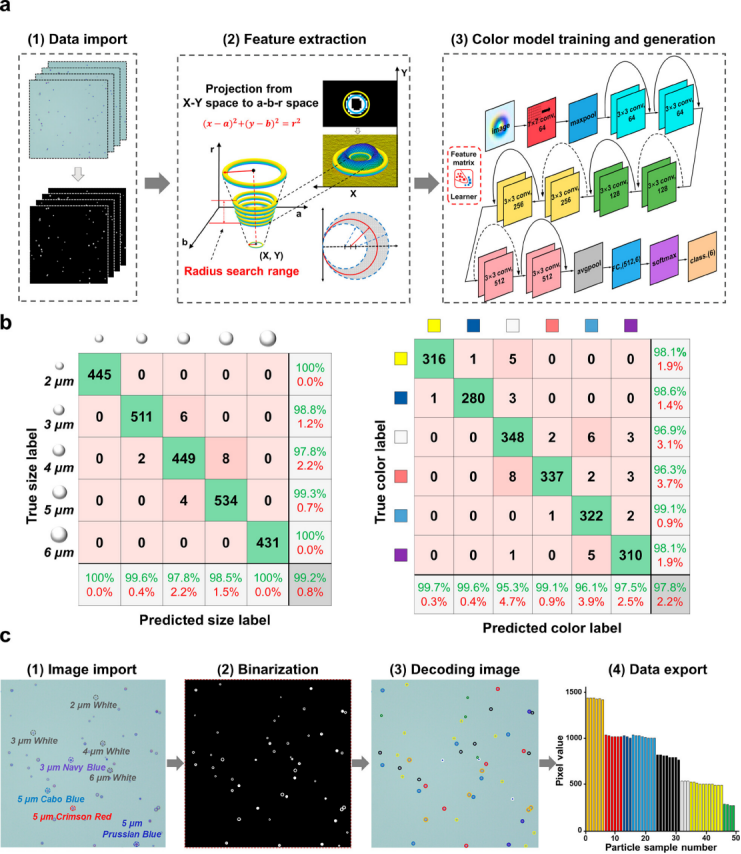
Figure 2 Principle of CAT.
3. Recognition Accuracy of CAT for Information Decoding
CAT was compared with Image-Pro Plus and RGB mode, testing images containing various encoded microspheres. Image-Pro Plus can recognize total counts and colors but cannot classify sizes, while RGB mode does not support quantitative and color classification. CAT can accurately recognize the quantity, color, and size of encoded microspheres. The accuracy of CAT counting was consistent with manual counting for three types of encoded microspheres and mixtures. A comparison of 35 images showed excellent consistency between CAT and manual decoding, with CAT recognition being faster; manual counting took over 3 minutes per image, while CAT took only 0.1 seconds, and CAT has high throughput and parallel processing capabilities.
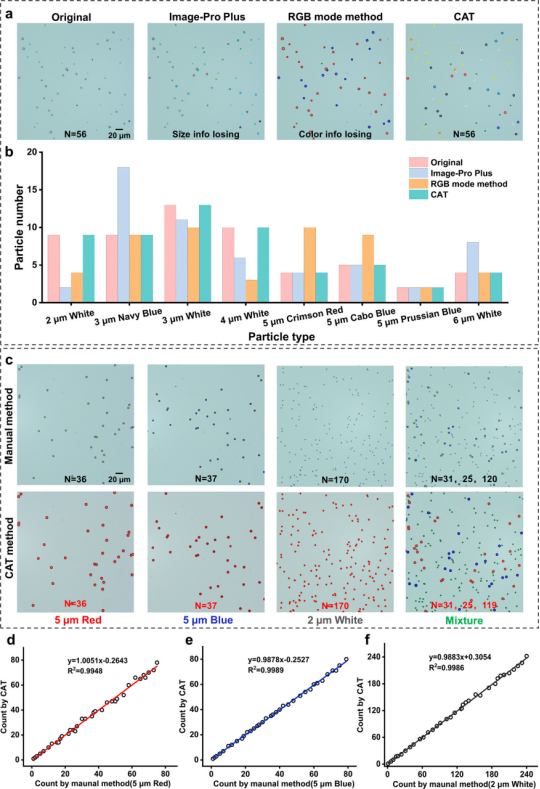
Figure 3 Recognition accuracy of CAT.
4. Sorting Theory and Mechanism of Microfluidic Chip
The microfluidic chip was designed to integrate reaction areas, signal sorting areas, and image acquisition areas. Particles in the magnetic sorting area are subjected to magnetic forces, gravity, and other forces, with the sorting mechanism based on magnetophoresis and fluid dynamics constraints. The magnetic force comes from magnetic fields in the X and Y directions, and relevant force calculation formulas are provided. Besides magnetic force, fluid dynamics constraints are important; fluid pressure and flow rate are analyzed based on Bernoulli’s principle, simulating flow rates in different channel cross-sections to analyze the velocity field. When PS microspheres do not bind with MNPs, they move along the main channel, while the immunocomplex moves to the side channel under the magnetic field, ensuring its flow rate to the image acquisition area. The chip reaches a stable state within 0.5 seconds, and fluid dynamics constraints are related to Reynolds number. PS4000 was selected for model validation of flow rate; when the flow rate is between 0.5 – 1.5 μL/min, it is feasible, above 1.8 μL/min is unrestricted, and below 0.3 μL/min makes it difficult for microspheres to reach the acquisition area. The mixing effect of curved reaction channels was studied using dye solutions, and imaging of the immunocomplex after treatment showed consistent results between simulation and experiments, indicating the feasibility of this strategy.
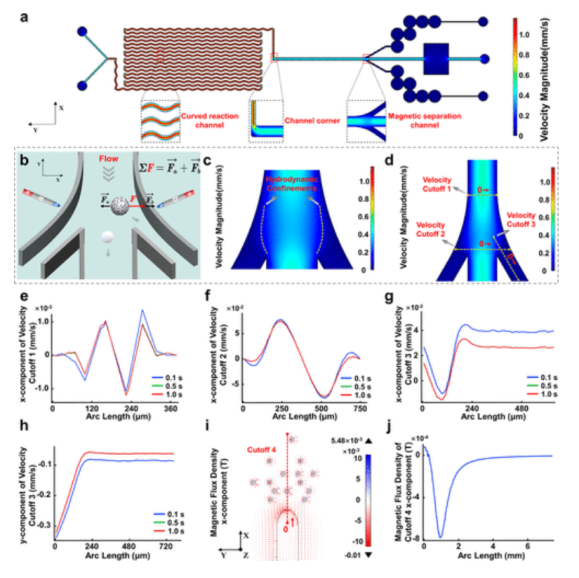
Figure 4 Simulation and analysis of physical fields in microfluidic chips for sorting signal probes using finite element method.
5. Multiplex Detection of Inflammatory Biomarkers
Detecting multiple biomarkers is beneficial for early diagnosis of diseases. Viral or bacterial infections can elevate CRP and IL-6 levels, while only bacterial infections increase PCT levels. Multiplex detection can provide comprehensive information. The CAT immunoassay method was used for multiplex detection of three inflammatory biomarkers, selecting red PS5000, blue PS5000, and white PS2000 to detect CRP, PCT, and IL-6, with small-sized encoded microspheres chosen for IL-6 detection due to its high detection requirements. After antibody coupling, the zeta potential of PSs changed, indicating good dispersion. SEM images show MNP150 on the surface of encoded microspheres, which are difficult to visualize with optical microscopy, reducing background interference. The quantity of encoded microspheres increased with the concentration of biomarkers, proving the feasibility of digital immunoassays.
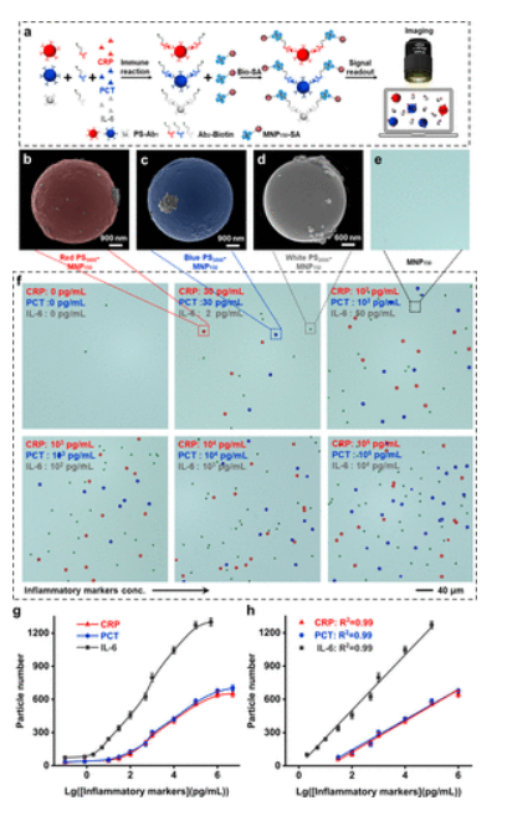
Figure 5 CAT immunoassay for detecting multiple inflammatory biomarkers.
After optimizing experimental conditions, three inflammatory biomarkers can be detected within specific linear ranges, with LODs of 2.39 pg/mL for CRP, 2.12 pg/mL for PCT, and 0.48 pg/mL for IL-6, showing advantages in sensitivity and accuracy compared to CLIA, with detection time reduced from 90 minutes to 30 minutes. Specificity was assessed, and the influence of interfering proteins was negligible. Detection of spiked serum samples showed good recovery rates, with RSD maintained below 15%. Analysis of patient samples showed good consistency between CAT immunoassay and CLIA or ELISA, with CRP detection covering clinical reference ranges. CAT immunoassay outperformed traditional methods in diagnosing infectious diseases and assessing cardiovascular disease risk.
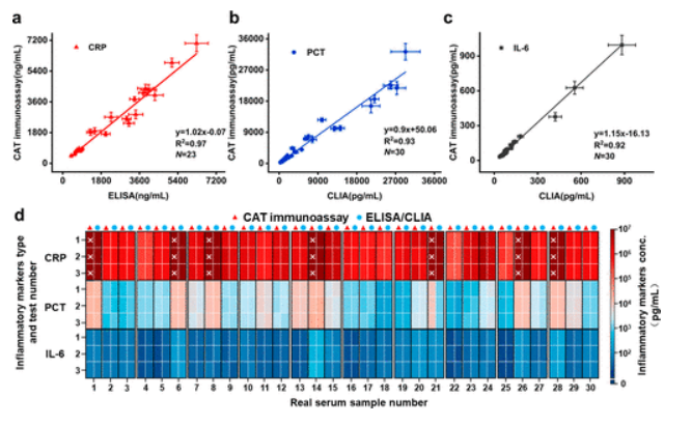
Figure 6 CAT immunoassay for detecting three inflammatory biomarkers in real serum samples.
6. Multiplex Detection of Three Antibiotics
To evaluate the multifunctionality of digital immunoassays, the CAT immunoassay method was used to detect three antibiotics: kanamycin, neomycin, and chloramphenicol. Different types of encoded microspheres were used as signal probes, and competitive immunoassays were conducted to detect the three antibiotics and PS-MNP150 complexes for quantification.
The performance of the CAT immunoassay method for detecting multiple antibiotics was evaluated, with linear ranges and LODs as follows: Kana 10 pg/mL to 100 ng/mL, 1.36 pg/mL; Neo 50 pg/mL to 500 ng/mL, 2.58 pg/mL; Cap 10 pg/mL to 100 ng/mL, 1.02 pg/mL. Compared to CLIA, the sensitivity of CAT immunoassay improved by 35 times, with a broader linear range. Accuracy was assessed by spiking milk samples, showing reasonable recovery rates with RSD maintained below 11%. Specificity in complex samples indicated good stability and specificity for small molecule detection. In real milk samples, multiple antibiotics were detected, with results comparable to CLIA validation. The CAT immunoassay showed good performance for analyzing antibiotics and biomarkers in mixed samples, indicating great potential for this low-cost, adaptable platform.
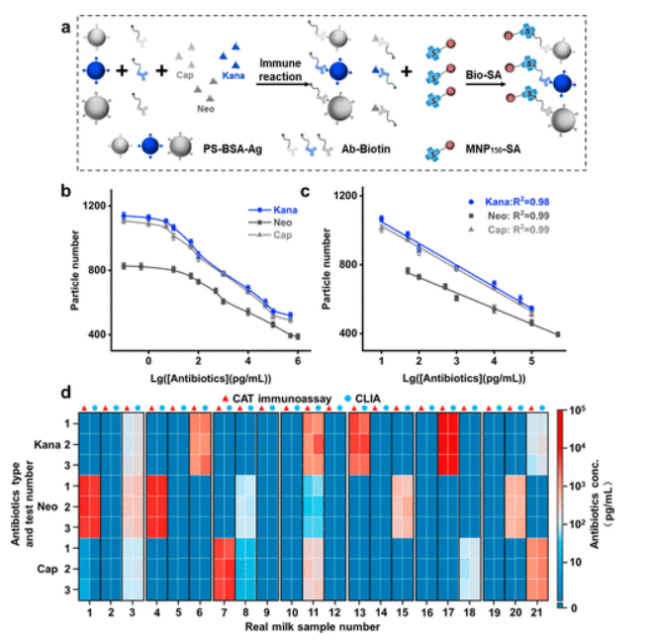
Figure 7 CAT immunoassay for detecting multiple antibiotics.
 Conclusion
Conclusion
This paper successfully developed a novel label-free surface-enhanced Raman scattering (SERS) spectral integrated microfluidic platform, combined with deep learning technology, for efficient detection and classification of exosomes derived from non-small cell lung cancer (NSCLC) cells and normal lung epithelial cells. This platform utilizes gold nanoclusters (AuNCs) functionalized with anti-CD-9 antibodies in combination with polystyrene (PS) microspheres to form capture probes with SERS activity for the capture and detection of exosomes. Within the microfluidic chip, this capture probe achieved an excellent capture rate of up to 85% and provided high-quality exosome SERS datasets. Using the obtained exosome SERS characteristic patterns, the deep learning model distinguished different NSCLC subtypes, achieving a classification accuracy of 97.88% and an area under the curve (AUC) value of 0.99 across all categories. Meanwhile, the gradient-weighted class activation mapping (Grad-CAM) technique highlighted interpretable molecular features in the spectral data to reflect the logical decision-making information behind the neural network.
This study demonstrates the application of combining SERS-coupled microfluidic technology with deep learning in liquid biopsy of exosomes, characterized by short analysis time, low sample consumption, high detection sensitivity, and powerful diagnostic capabilities. The results are expected to significantly improve the early diagnosis rate of NSCLC, precise subtype classification, and personalized treatment outcomes.
Comments1. This paper proposes a computer vision-based AI technology for multidimensional encoding and decoding in digital immunoassays. By combining particle encoding and AI decoding, it achieves high sensitivity, high specificity, and simultaneous detection of multiple biomarkers.2. The study utilizes the size, quantity, and color characteristics of microspheres for information encoding and decodes microscopic images using computer vision algorithms, enabling rapid and accurate identification and quantitative analysis of multiple targets.3. The study combines computer vision technology with AI algorithms to achieve automatic decoding and analysis of microscopic images. This combination not only improves detection efficiency but also reduces human operational errors, bringing new technical means to the field of biosensing.
 Original Link
Original Link
https://pubs.acs.org/doi/10.1021/acsnano.3c02941
FOLLOW US
For submissions, recommendations, and collaborations, please contact us!
|| WeChat Official Account: Micro-Nano Sensing
|| WeChat ID: successplus999
Micro-Nano Sensor Industry Research
Academic Frontier Research Reports

Author: Ding Chenchen
Proofreader: Teacher Zhang
Publisher: Tian Tiantian
Disclaimer ————
This article is for research sharing only and not for profit. If there is any infringement, please contact the backend staff for deletion.
Due to limited knowledge, there may be omissions and errors; please feel free to criticize and correct.