Hello everyone, today I would like to share an article published in JACS: After 75 Years, an Alternative to Edman Degradation: A Mechanistic and Efficiency Study of a Base-Induced Method for N‑Terminal Peptide Sequencing. The corresponding authors are Professors Eric V. Anslyn and Edward M. Marcotte from the University of Texas at Austin. Professor Anslyn’s research group mainly focuses on physical organic chemistry, supramolecular chemistry, and materials chemistry, and designs various sensing systems; Professor Marcotte’s research interests include protein function, expression, and localization in proteomics, as well as single-molecule protein sequencing technologies.

Edman degradation is a classic peptide sequencing technique that involves the repeated use of phenylisothiocyanate (PITC) to treat the N-terminal amino group of peptides, followed by treatment with trifluoroacetic acid to obtain the amino acid sequence information of the peptide chain. It has played an important role in biochemistry and molecular biology. However, with the development of proteomics, the demands for protein sequencing technologies have increased, and traditional Edman degradation is limited in certain cases, such as when dealing with acid-sensitive peptide segments or proteins containing acid-sensitive functional groups. Therefore, developing a new non-acidic N-terminal peptide sequencing method is of significant scientific and practical value.
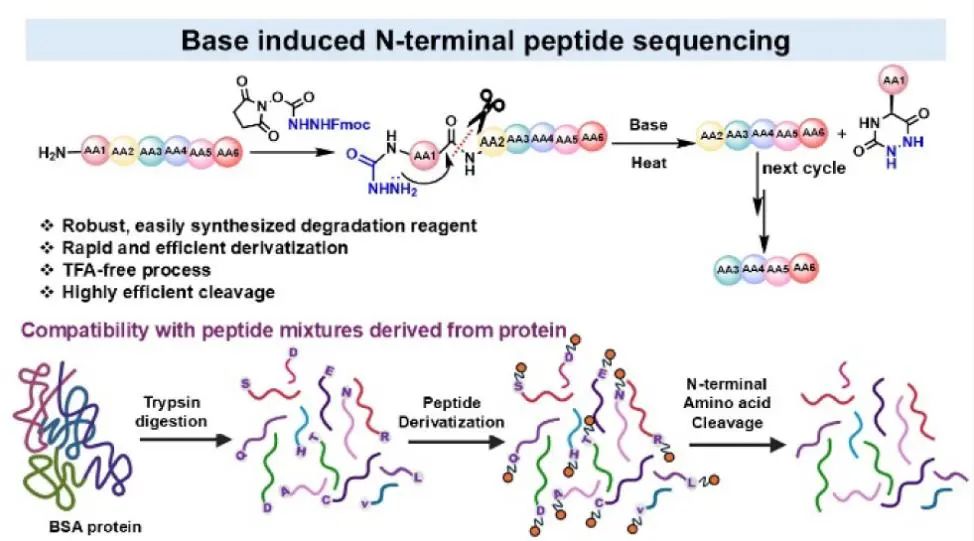
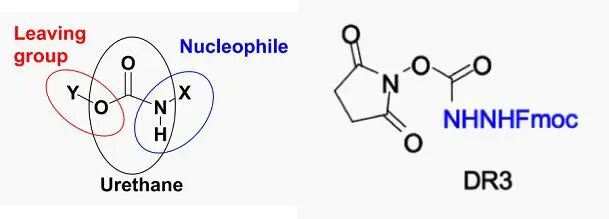
The main goal of the authors is to develop a base-induced degradation method. According to literature reports, in non-enzymatic catalysis, bases can assist in the hydrolysis of amide bonds, with a mechanism involving nucleophilic attack of hydroxide on the carbonyl carbon, forming a tetrahedral intermediate. The second step of degradation involves the cyclization of the peptide and the departure of the amino group. The authors studied various peptide cleavage cyclization reactions, for example, under high temperature (130 °C) and neutral pH conditions, the intramolecular cyclization between the N-terminal amino acid and the adjacent amide can form diketopiperazine and release truncated peptides. Inspired by the literature, the authors iteratively improved the performance of degradation reagents (DRs) by altering leaving groups and nucleophilic species, with two key components: one is the leaving group for selective N-terminal derivatization, and the other is a good nucleophile to promote cyclization. The authors selected N-hydroxysuccinimide as the leaving group and hydrazine formamide as the super nucleophile to synthesize various N-terminal derivatization reagents. After multiple rounds of iterative design and experimental screening, they found that compound DR3 yielded the highest efficiency in the peptide derivatization step and could effectively eliminate the N-terminal amino acid with only 1% hydroxide salt.
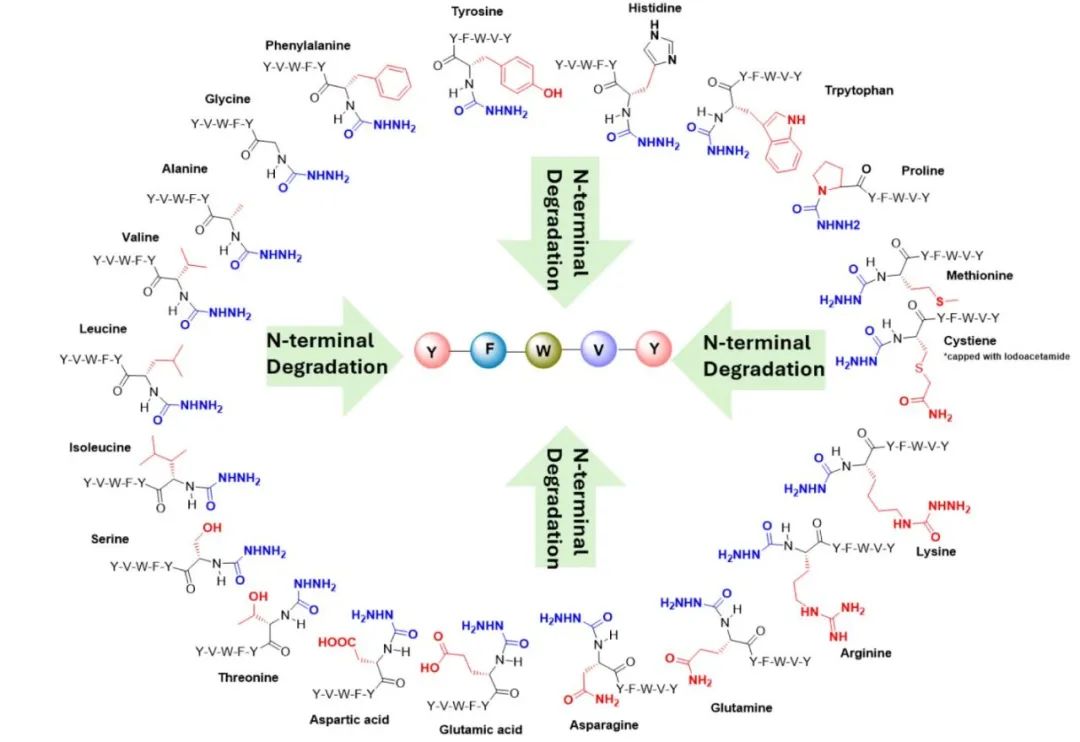
Subsequently, the authors reacted the peptide with DR3 under specific conditions, allowing the N-terminal amino acid to undergo derivatization with DR3. Then, a base was added to promote the elimination of the N-terminal amino acid. The reaction products were analyzed using techniques such as LC-MS/MS to determine the efficiency of the derivatization and elimination reactions, and to examine the applicability of this method to different amino acids. The authors successfully removed all 20 amino acids from the N-terminus using this method, and compared to Edman degradation, it exhibited higher sequencing efficiency and accuracy for certain specific amino acids (such as cysteine and methionine), which may be due to the higher reactivity or stability of these amino acids under basic conditions.
In summary, the authors have developed a new base-induced N-terminal degradation method, providing a valuable alternative for protein sequencing, especially in handling acid-sensitive samples and in cases where simultaneous analysis of proteins and nucleic acids is required.
Author: MB
Editor: LZ
DOI: 10.1021/jacs.5c03385
Original link: https://doi.org/10.1021/jacs.5c03385
