“Natural selection only acts on the minute, continuous variations.”
—— Charles Darwin, On the Origin of Species
Throughout the history of species evolution, the expansion of brain capacity in modern humans (Homo sapiens) is one of the most remarkable achievements. Approximately 6-7 million years ago (ma), humans diverged from their closest ape relatives, the chimpanzees (Pan troglodytes), beginning their own independent evolutionary branch【1】. The DNA similarity in the protein-coding regions of the genomes of humans and chimpanzees is about 99.1%, which means that the genetic code determining the participation in the “central dogma” cannot fully explain the differences between humans and apes. In 1975, King and Wilson proposed that the evolutionary mysteries of humans and chimpanzees lie in the differences in gene expression regulation【2】.
Human Accelerated Regions (HARs) are highly conserved DNA sequences widely distributed in mammalian genomes; however, these DNA fragments exhibit accelerated accumulation of fixed nucleotide substitutions after the divergence of the human lineage. Since HARs are located very close to genes related to neurodevelopment on the chromatin, researchers have been attempting to find answers regarding human brain evolution from these sequences for the past 20 years. Currently, it is known that there are approximately 3000 HAR fragments distributed across various chromosomes in the human genome, with an average length of about 300 bp, and about 50% of HARs exist as enhancers in the nervous system【3】【4】, participating in neuronal maturation【5】. However, the biological mechanisms by which HAR sequences regulate the expansion of human brain capacity remain unknown.
On May 14, 2025, a team led by Debby Silver at Duke University published a research paper titled A human-specific enhancer fine-tunes radial glia potency and corticogenesis in Nature, which for the first time deeply analyzed the biological functions of human accelerated regions (HARs) in the process of cortical development using cross-species models of mice, chimpanzees, and humans.

The researchers analyzed data from human brain HiC, DNaseI-seq, and H3K27ac ChIP-seq to preliminarily identify a human accelerated region enhancer, HARE5 (HAR enhancer No.5), from the existing HAR sequence library. HARE5 (also known as ANC516; HARsv2_0258) is located on human chromosome 10 (Hg38chr10:35949493-35950111), with a total length of 619 bp containing four human-specific nucleotide substitutions. HARE5 acts as a highly active enhancer in human neural stem cells, directly regulating the transcription level of the target gene FZD8 (Fizzled8). FZD8, as one of the receptors in the WNT signaling pathway, is specifically expressed on the surface of neural stem cells in the brain cortex of humans, macaques, and mice (Radial Glia Cells, RGCs). To further explore the biological function of HARE5, the researchers replaced the sequence in the mouse genome (Mm-HARE5) with the human-derived sequence (Hs-HARE5), resulting in a knock-in of the human HARE5 mouse model. Analysis showed that the human HARE5 adult mice exhibited a 5% expansion in cortical area and a significant increase in mature excitatory neurons. Functional calcium imaging results indicated that the increase in brain volume promotes the independence of functional brain regions, specifically manifested as weakened connectivity between brain regions.
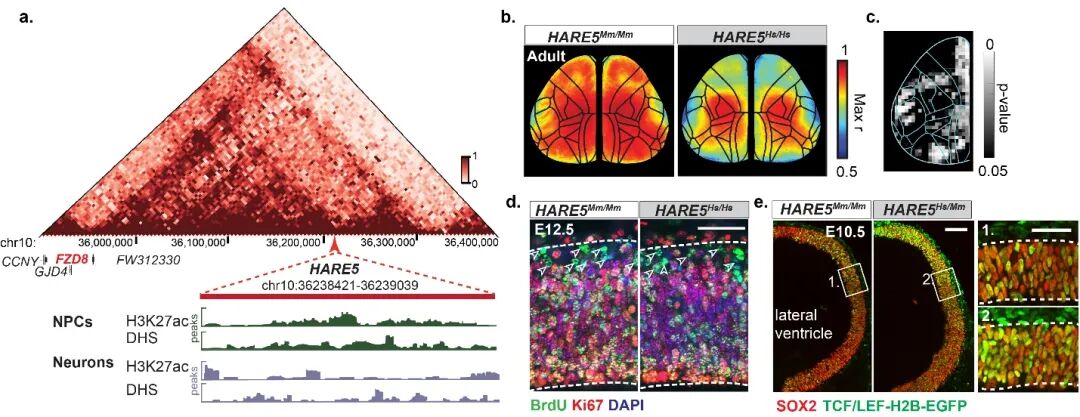
Figure 1. HARE5 fine-tunes mouse cortical construction and neural connectivity.
The changes in brain volume primarily stem from differences in cortical construction during embryonic development. Therefore, to further explore the differences and regulatory mechanisms of cortical development between humans and chimpanzees, the researchers employed CRISPR-Cas9 to perform knock-in genome editing on human embryonic stem cell lines (ESCs, H9), replacing the Hs-HARE5 sequence with the chimpanzee sequence (Pt-HARE5) and inducing them into cortical organoids. Although there were no significant morphological and structural differences between the human and chimpanzee-derived organoids, further analysis revealed that human neural stem cells carrying Pt-HARE5 exhibited a significant reduction in the number and proliferation capacity of neural stem cells due to the low activity of the enhancer, leading to a weakening of the canonical WNT signaling pathway. Additionally, experiments detecting the WNT reporter in embryonic human HARE5 mice and RNA sequencing of the cortical tissue at different stages confirmed that HARE5 influences cortical construction by regulating the levels of canonical WNT signaling in early neural stem cells (RGCs). Meanwhile, the researchers also performed knock-in genome editing on chimpanzee induced pluripotent stem cell lines (iPSCs, C3649), replacing the Pt-HARE5 sequence with the human sequence (Hs-HARE5) and inducing them into cortical neural stem cells. Cell cycle analysis showed that the proliferation capacity of chimpanzee cortical neural stem cells carrying Hs-HARE5 significantly increased. Notably, the regulation of ancient signaling pathways (such as FGF-ERK[6]; mTOR[7]) is increasingly recognized as an important mechanism in the evolution of the human brain.
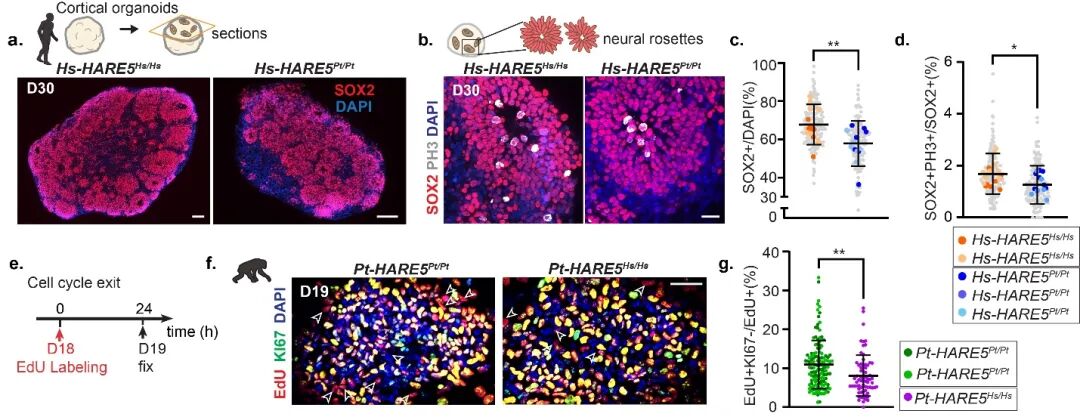
Figure 2. HARE5 sequence differences lead to differences in the proliferation capacity of human and chimpanzee neural stem cells.
In summary, this work, through rich research methods and species models, has for the first time identified the cellular and molecular mechanisms by which HARs in the human genome directly regulate the expansion of human brain capacity. This research provides new evidence for understanding how small genomic sequence changes drive the unique evolution of the human brain in the primate lineage, and opens new perspectives and directions for understanding the complex regulation of brain development and pathogenic processes.
Millions of years of natural selection have accumulated complex and diverse variations in the human genome, including single nucleotide polymorphisms (SNPs), insertions or deletions of DNA fragments (INDELs), segment duplications (SDs), copy number variations (CNVs), and more. These intrinsic driving forces within the genome are like the spark of fire brought to humanity by Prometheus, allowing us, as pilgrims through the long ages, to develop opposable thumbs, learn to walk upright, and create language and civilization. Gazing at the vast universe is not only to look up at the starry sky above but also to introspect the myriad stars within our genome. The biological functions of HARE5 deepen our understanding of the role of HARs in species evolution, but ultimately, it is just the tip of the iceberg in the study of non-coding sequence regulatory elements and brain evolution. Many scientific questions remain to be explored, such as how HARs participate in the evolution of modern humans and ancient humans; how regulatory sequences co-evolve with cell-specific transcription factors; how the human chromatin-specific fine spatial conformations (TAD, Loop) achieve the biological functions of cortical development【8】; and how different regulatory elements (cis and trans) complete multi-level regulation.
Dr. Jing Liu from Duke University, the first author of this article, and Professor Debby Silver, the corresponding author, warmly welcome scholars with a strong interest in exploring the mechanisms of human brain evolution to join us.
Original link:
https://doi.org/10.1038/s41586-025-09002-1
Editor: Eleven
Source: BioArt
References
1. Pollen, A.A., et al., Human-specific genetics: new tools to explore the molecular and cellular basis of human evolution. Nature Reviews Genetics, 2023. 24(10): p. 687-711.
2. King, M.-C. and A.C. Wilson, Evolution at Two Levels in Humans and Chimpanzees. Science, 1975. 188(4184): p. 107-116.
3. Lindhout, F.W., et al., A molecular and cellular perspective on human brain evolution and tempo. Nature, 2024. 630(8017): p. 596-608.
4. Girskis, K.M., et al., Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron, 2021. 109(20): p. 3239-3251.e7.
5. Cui, X., et al., Comparative characterization of human accelerated regions in neurons. Nature, 2025. 640(8060): p. 991-999.
6. Zhang, Z., et al., ERK and PKA signaling drive the evolutionary expansion of the cortex. bioRxiv, 2025: p. 2025.04.09.647935.
7. Andrews, M.G., L. Subramanian, and A.R. Kriegstein, mTOR signaling regulates the morphology and migration of outer radial glia in developing human cortex. eLife, 2020. 9: p. e58737.
8. Luo, X., et al., 3D Genome of macaque fetal brain reveals evolutionary innovations during primate corticogenesis. Cell, 2021. 184(3): p. 723-740.e21.


Scan to follow us
WeChat ID丨sscbweixin
Website丨https://sscb.org.cn/