Hello everyone, today I am sharing an article published in JACS titled “Multiplicity of Regulatory Subunit Conformations Defines Structural Ensemble of Reset Protein Kinase A Holoenzyme.” The corresponding author is Professor Ganesh S. Anand from Pennsylvania State University, whose research focuses on using mass spectrometry tools to reveal the dynamic processes of cellular macromolecular assembly.

As is well known, Protein Kinase A (PKA) holoenzyme consists of two catalytic subunits (C) and two regulatory subunits (R). In the inactive state, the pseudo-substrate sequence of the R subunit binds to the catalytic site of the C subunit; when the two cAMP binding domains (CNB-A/B) of the R subunit bind cAMP, the R subunit undergoes a conformational change and dissociates from the C subunit, thereby activating the kinase activity of PKA. However, how the cAMP-modulated R subunit is reset and how it rebinds to the C subunit is not fully understood. Current evidence suggests that phosphodiesterases (PDEs) anchored with PKA on A-kinase anchoring proteins (AKAPs) participate in the hydrolysis of cAMP, which is involved in the resetting of PKA-R, but the dynamic changes in protein structure during this process remain unknown. In this article, the authors used tools such as cryo-electron microscopy (cryo-EM), negative stain electron microscopy (nsEM), native mass spectrometry (native MS), hydrogen-deuterium exchange mass spectrometry (HDXMS), and cross-linking mass spectrometry (XLMS) to characterize the conformational landscape during the PDE8-mediated resetting process of the PKA holoenzyme in detail.
First, the authors determined through native MS that PDE8 is essential for the resetting of PKA: PKA-RIα saturated with cAMP can no longer re-combine with PKA-Cα, while after incubation with PDE8, the reformation of the holoenzyme can be observed, and this resetting process partially depends on ATP.
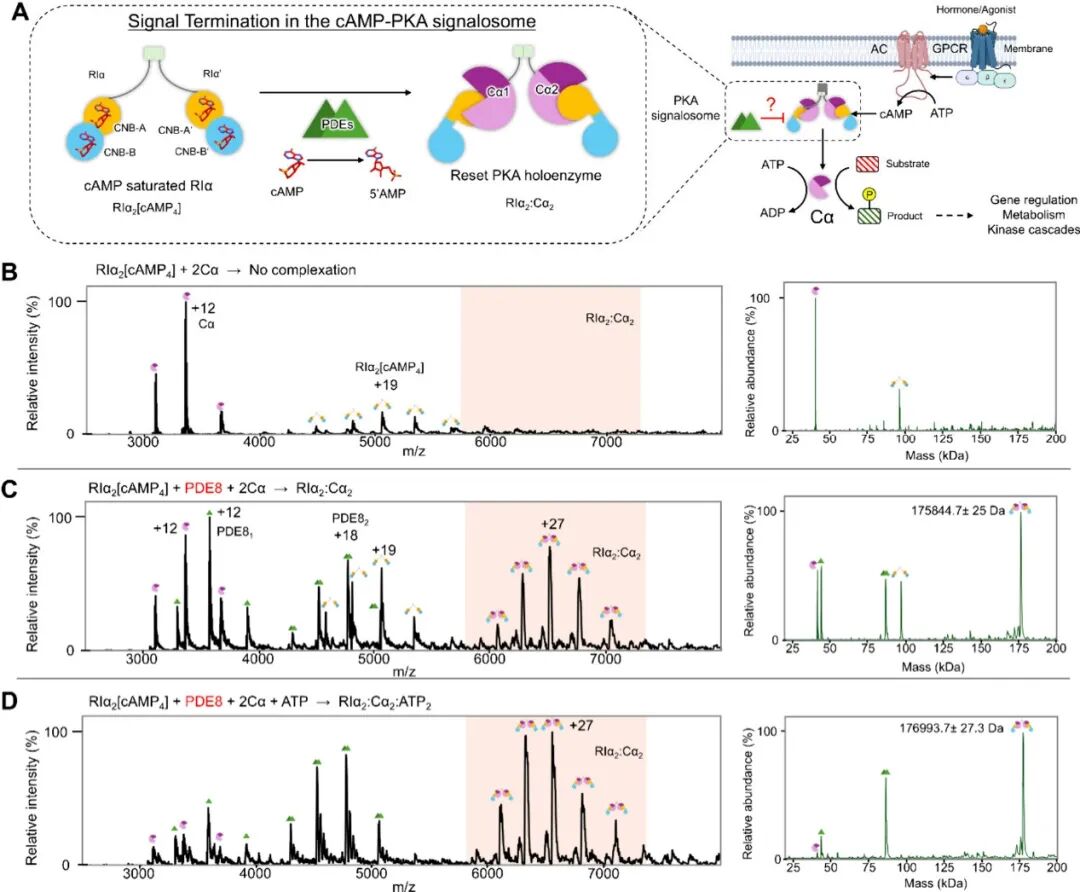
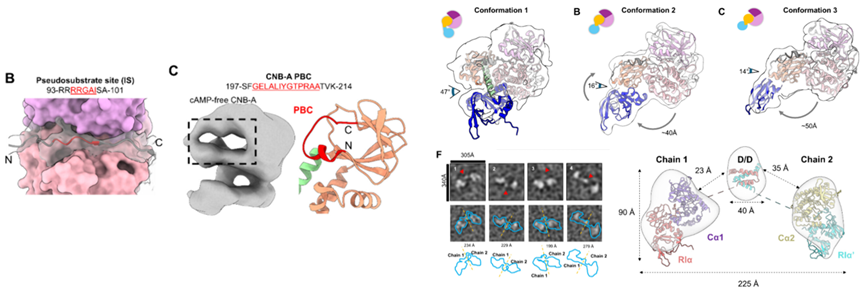
Subsequently, the authors used cryo-EM and HDXMS to resolve the structure of the PKA holoenzyme after resetting, revealing that the reset PKA holoenzyme is a highly dynamic heterotetramer, where the CNB-A of RIα forms a stable interface (binding ATP) with Cα, while CNB-B is highly disordered and no density was observed. HDXMS confirmed that cAMP is completely released from CNB-A/B. Further structural analysis found that CNB-B exists in three spatial conformations (with a 40Å to 14° angle variation), and its dynamics are driven by the breakage of the αB/C helix connecting CNB-A/B, with HDXMS showing enhanced hydrogen bond protection in specific regions after cAMP release from CNB-B. Additionally, HDXMS and XLMS revealed the dynamic characteristics of the disordered linker region at the N-terminus of RIα (I63-D119), where the pseudo-substrate sequence is well stabilized by the bound Cα. The C-terminal linker region (111-123) is highly dynamic. Negative stain electron microscopy showed that the holoenzyme particles are dumbbell-shaped, with a wide distribution of inter-chain distances (160-320Å), reflecting high flexibility between the chains.
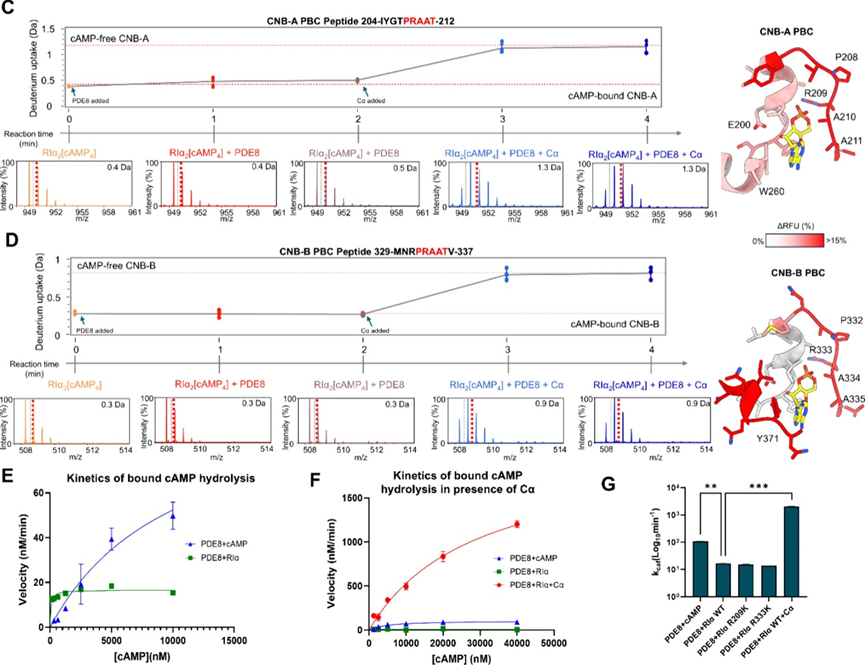
Finally, the authors aimed to time-resolve the dynamic process of PDE8 hydrolyzing cAMP into 5’AMP and mediating the formation of the PKA holoenzyme. They used pulse-labeling HDXMS analysis to monitor the release of cAMP from RIα, sampling every minute over a 4-minute time scale. The results showed that when PDE8 acts alone on PKA-RIα, the hydrolysis rate of bound cAMP is slow (8 times slower than free cAMP). Interestingly, in the presence of PKA-Cα, the hydrolysis rate of bound cAMP increases by about 130 times, and the resetting of the holoenzyme can be completed within one minute. These results reveal that the resetting of PKA is achieved through a PDE8 cooperative mechanism assisted by Cα, where Cα accelerates cAMP dissociation, and PDE8 completes hydrolysis to drive holoenzyme formation.
In summary, this article reports the degradation of cAMP bound to PKA-RIα mediated by PDE8 and the resetting of the PKA holoenzyme, using various tools to detail the conformational landscape during the resetting process of the PKA holoenzyme, including the stable interface at CNB-A, the conformational diversity of CNB-B, and the dynamics of the linker regions. The authors also preliminarily explored the mechanism of Cα-assisted cAMP dissociation and hydrolysis.
Author: TYC
Editor: MB
DOI: 10.1021/jacs.4c16269
Original link: https://doi.org/10.1021/jacs.4c16269
