Scientific Review



Click the link below to read the original article

Commentators
Zhong Jianli* and Fang Xuanjun
Hainan Tropical Agricultural Resources Development and Utilization Research Institute, Sanya, 572024
*Contact Email, [email protected]

On March 13, 2024, a collaborative research result from the State Key Laboratory of Subtropical Agricultural Biological Resources Protection and Utilization at South China Agricultural University, the Guangdong Academy of Agricultural Sciences Rice Research Institute, and the Life Science Technology Center of China Seed Group Co., Ltd. was published in the journal Nature Communications. Guo Yajun and Heng Yueqin are co-first authors, while Shen Rongxin and Wang Haiyang are co-corresponding authors, titled “Natural variation in OsMYB8 confers diurnal floret opening time divergence between indica and japonica subspecies”. This study was supported by the National Natural Science Foundation’s Innovation Group Project, the Hainan Yazhou Bay Seed Laboratory’s leadership project, and others. The research indicates that the OsMYB8 gene is a key factor regulating the differences in diurnal flowering time in rice. The identified interaction between OsMYB8 and OsJAR1 significantly affects the flowering time of indica and japonica rice. Transferring the indica OsMYB8 allele into japonica rice can effectively advance flowering time, providing a new strategy for indica-japonica hybrid breeding.
1 Experimental Data Analysis
This study meticulously recorded the flowering behavior of indica and japonica rice at different times of the day. RNA sequencing (RNA-seq) analysis revealed that the expression level of OsMYB8 increases before flowering, and this upregulation acts by directly promoting the expression of the OsJAR1 gene. The interaction between OsMYB8 and OsJAR1 further influences the expression of genes related to regulating cell permeability and cell wall remodeling, collectively promoting flower opening.
Figure 1 shows the differences in diurnal flower opening time (DFOT) between the two rice subspecies. Figure 1a compares the DFOT of indica (n=28) and japonica (n=12) rice varieties in Guangzhou in October 2019, showing that indica varieties generally flower earlier than japonica varieties. Figures 1b and 1c compare the flowering of indica rice TFB and japonica ZH11 at 10:30 AM and 12:00 PM, with significant differences, where TFB flowers open earlier. Figures 1d and 1e record the changes in DFOT of TFB and ZH11 in June and October 2020, indicating the stability of DFOT in different seasons. Figure 1f presents stereomicroscope photos showing the changes in pistil morphology of TFB and ZH11 at different times of the day, while Figure 1g illustrates how to measure the volume of the pistil as an ellipsoid. Figure 1h quantitatively compares the pistil volumes of ZH11 and TFB at different time points, calculated using the ellipsoid volume formula. Finally, Figure 1i analyzes the water content of 100 pistils of ZH11 and TFB at different time points, providing experimental data for further exploring the relationship between DFOT and water content.
Thus, these results highlight the significant differences in DFOT between the two rice subspecies, which is of great importance for rice breeding and understanding the impact of different biological clocks on plant flowering.
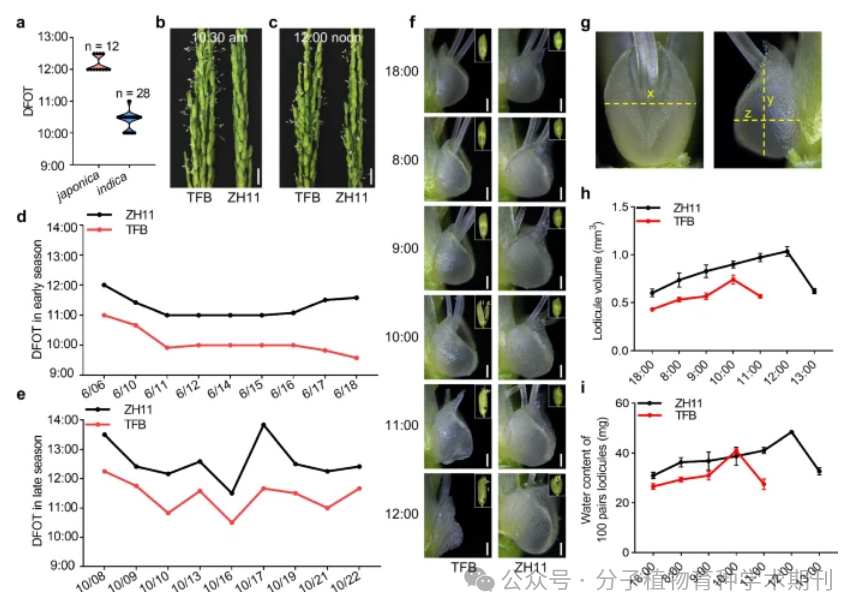
Figure 1 Differences in diurnal flower opening time (DFOT) among rice subspecies
Figure 1 DFOT divergence in rice subspecies
Figure 2 reveals the gene expression differences in the pistils of indica (TFB) and japonica (ZH11) rice at different time points through comparative transcriptomic analysis. Figure 2a is a principal component analysis (PCA) showing the differences in the transcriptomic datasets of TFB and ZH11 pistils at different time points, where time points T18 and Z18 represent 18:00 the day before flowering, T9 and Z9 represent 9:00 before flowering (corresponding to 1 hour and 3 hours before flowering for TFB and ZH11, respectively), and Z11 represents 11:00 (1 hour before ZH11 flowers), while TF and ZF represent the peak flowering times (approximately 10:00 for TFB and 12:00 for ZH11). The Venn diagram in Figure 2b shows the intersection of differentially expressed genes (DEGs) upregulated in Cluster18 genes compared to T9 and Z9. Figure 2c is a gene ontology (GO) enrichment analysis of the 251 intersecting genes, where molecular function (MF) and biological process (BP) are marked in different colors, indicating different levels of P values, with “transcription factor activity” highlighted in red. Figure 2d shows the expression analysis of 11 transcription factors selected through GO analysis, with numbers in the heatmap representing average FPKM values. Figure 2e presents reverse transcription quantitative PCR (RT-qPCR) analysis of the expression levels of OsMYB8 in TFB and ZH11 pistils at 9:00 AM in June and October 2021, with values representing mean ± standard error (SEM), n=3 biological replicates, and significance assessed using a two-tailed Student’s t-test, providing P values.
Thus, it indicates that there are significant differences in the expression regulation of pistils between indica and japonica rice, especially in the regulation of flowering time, where the OsMYB8 gene exhibits differential expression, which may be an important factor leading to the DFOT differences between the two subspecies.
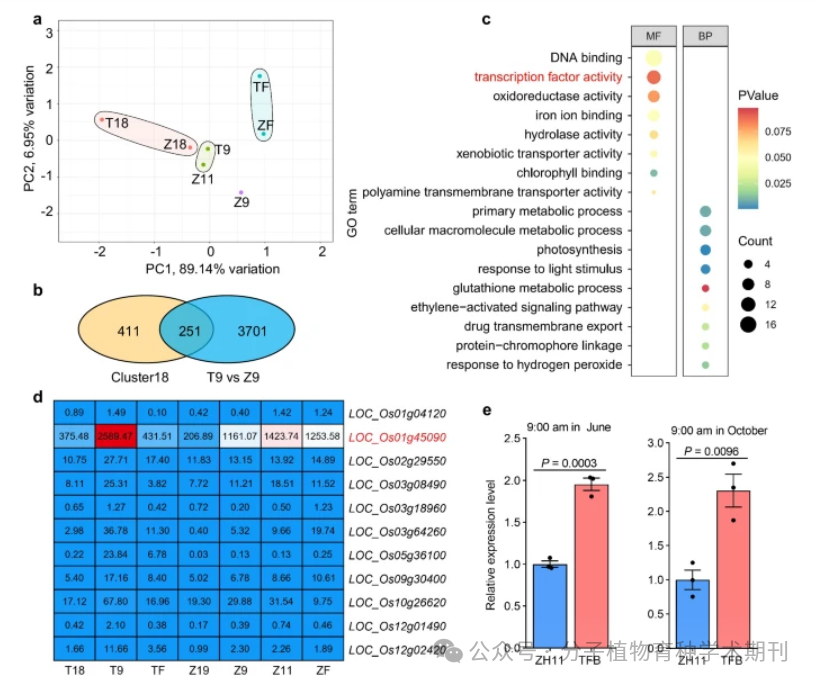
Figure 2 Identification of OsMYB8 by comparative transcriptomic analysis of lodicules between indica and japonica
Figure 3 demonstrates the use of CRISPR/Cas9 gene editing technology to create OsMYB8 gene mutants and analyze the effects of these mutants on diurnal flower opening time (DFOT) in different backgrounds (ZH11 and TFB). Figure 3a shows the successful introduction of OsMYB8 gene target mutations through CRISPR/Cas9 technology. Figures 3b, e, j compare the flowering of ZH11 and Osmyb8ZH mutants (b), TFB and Osmyb8TF mutants (e), and ZH11 and OsMYB8TF/ZH11 transgenic lines (j) at different time points. Figures 3c, f, k record the number of flowers opened by these plants at different times in June, while Figures 3d, g, l show the number of flowers opened after different days, both showing significant differences. Figure 3h is a schematic diagram of the vector structure used to construct OsMYB8TF/ZH11 transgenic plants. Figure 3i presents the expression levels of the OsMYB8 gene in the pistils of OsMYB8TF/ZH11 transgenic lines analyzed by RT-qPCR, with ZH11 as a negative control, showing a significant increase in the expression of OsMYB8 in the transgenic lines, determined to be significant through a two-tailed Student’s t-test.
Thus, it emphasizes the role of the OsMYB8 gene in positively regulating rice DFOT and indicates that it affects the number of flowers opened in the inflorescence, thereby influencing the reproductive efficiency of rice. In this way, researchers can better understand and manipulate key developmental stages in the growth process of rice.
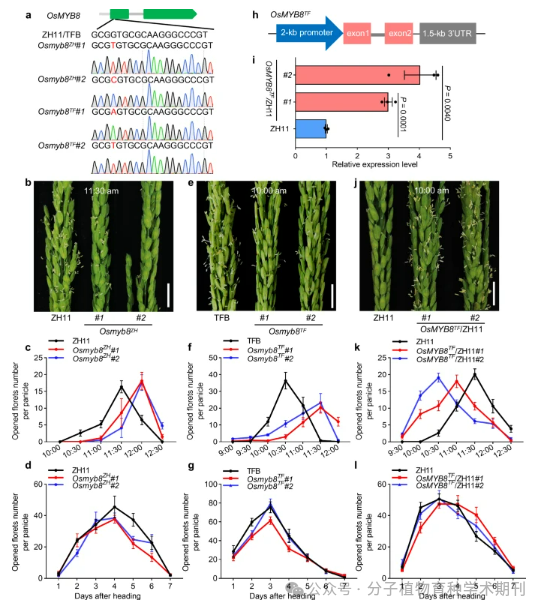
Figure 3 OsMYB8 positively regulates rice DFOT
Figure 4 presents the identification process of the genome-wide direct target genes of OsMYB8 and their functional analysis. a) The nuclear localization of OsMYB8-GFP in rice protoplasts was visualized by co-localization with D53-mCherry nuclear marker. b) Yeast two-hybrid experiments show that both full-length and truncated OsMYB8 proteins fused with the DNA binding domain (BD) have transcriptional activation activity. c) The core binding motif “TTHGGY” of OsMYB8 protein was identified using the MEME-ChIP tool. d) The Venn diagram compares the genes bound by DAP-seq technology with the differentially expressed genes (DEGs) identified in the Osmyb8 mutant through RNA-seq. e) GO (gene ontology) enrichment analysis of 345 overlapping genes, where MF represents molecular function, BP represents biological process, and the depth of color indicates the size of P values, with the size of circles representing the number of genes in the corresponding functional category. f) ChIP-qPCR analysis indicates that OsMYB8 can bind to the promoter region of OsJAR1 in vivo. The red line marks the position of the “TTHGGY” motif in the OsJAR1 promoter region. g) Electrophoretic mobility shift assay (EMSA) shows that GST-OsMYB8 recombinant protein can directly bind to the OsJAR1 promoter region containing the “TTHGGY” motif. h) Transient dual-luciferase (LUC) reporter gene experiments indicate that OsMYB8 can induce the transcription of the OsJAR1 promoter in rice protoplasts. The LUC/REN ratio represents the relative activity of the OsJAR1 promoter.
Thus, the results further confirm the critical role of OsMYB8 in regulating diurnal flower opening time (DFOT) in rice, particularly its direct action on downstream target genes such as OsJAR1, thereby affecting JA-mediated signaling pathways and related biological processes. Additionally, these findings reveal the important role of transcription factors in plant growth and development and in response to environmental changes.
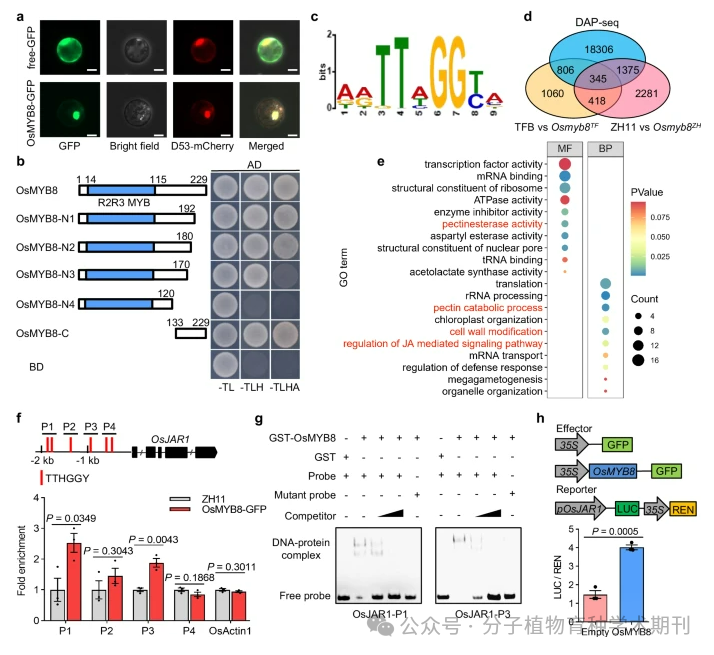
Figure 4 Identification of the genome-wide direct targets of OsMYB8
Figure 5 details the role of the OsJAR1 gene in regulating diurnal flower opening time (DFOT) in rice and its relationship with the content of jasmonic acid isoleucine (JA-Ile). a) The Osjar1 mutant was created in the ZH11 background using CRISPR/Cas9 gene editing technology, with the mutation site marked in red. b) It shows the comparison of spikes between ZH11 and Osjar1 mutants at 11:30 AM in Guangzhou in June 2022, with a scale of 1 cm. c) It records the number of flowers opened per spike of ZH11 and Osjar1 mutants at different times of the day, with averages represented as standard error (SEM) (n=10 spikes). d) and e) respectively measure the JA-Ile content in the pistils of ZH11, Osjar1 mutants, and Osmyb8ZH mutants at 10:00 AM and 9:00 AM, with values represented as standard error (SEM) (n=3 biological replicates). f) A schematic diagram shows the vector structure used to construct OsJAR1com materials, where pOsMYB8TF indicates that the promoter is amplified from TFB. g) It shows the relative expression levels of OsJAR1 in the pistils of ZH11, OsJAR1com, and Osmyb8ZH, with values represented as standard error (SEM) (n=3 biological replicates). h) and j) respectively show the comparison of spikes between ZH11, OsJAR1com, Osmyb8ZH, and ZH11, OsMYB8TF/ZH11, OsMYB8TF/Osjar1, and Osjar1 at 11:30 AM and 12:00 PM in Guangzhou in June and October 2022, with a scale of 1 cm. i) and k) respectively record the number of flowers opened at different times of the day for ZH11, OsJAR1com, Osmyb8ZH, and ZH11, OsMYB8TF/ZH11, OsMYB8TF/Osjar1, and Osjar1, with averages represented as standard error (SEM) (n=10 spikes).
Thus, the results indicate that OsJAR1 not only regulates rice DFOT by affecting JA-Ile content but also involves other complex biological processes. The data provided in the figures emphasize the important application of gene editing technology in functional gene research and the potential for regulating flowering time in rice breeding.
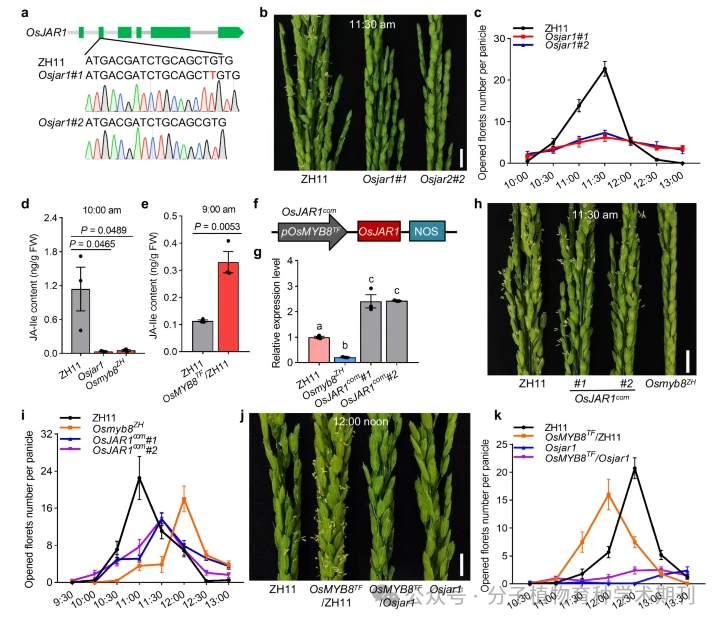
Figure 5 OsJAR1 influences JA-Ile content in lodicule to regulate rice DFOT
Figure 6 reveals how natural variation in the OsMYB8 promoter leads to differences in diurnal flower opening time (DFOT) between indica and japonica rice. a) A single nucleotide polymorphism analysis of the OsMYB8 promoter was conducted among 3513 rice germplasm resources, showing the variation in the 2 kb promoter region. b) It displays the distribution frequency of three OsMYB8 haplotypes in different Asian rice germplasm, with haplotype 1 (Hap1) being the most abundant. c) It analyzes the nucleotide diversity (π value) of the 100 kb genomic region surrounding the OsMYB8 gene, comparing the differences among indica (Ind), temperate japonica (TeJ), and wild rice (Ruf). d) The FST value reflects the level of genetic differentiation in the 100 kb region surrounding OsMYB8 among the three types mentioned above. e) It records the DFOT of rice germplasm containing Hap1 and Hap2 in Guangzhou in May 2022, finding that Hap1 germplasm generally flowers earlier than Hap2 germplasm. f) It measures the relative expression levels of OsMYB8 in the pistils of rice germplasm containing Hap1 and Hap2. g) It measures the JA-Ile content in the pistils of TFB and ZH11 at 9:00 AM. h) The transient dual-luciferase reporter gene experiment shows the transcriptional activity of pOsMYB8Hap1 and pOsMYB8Hap2 in rice protoplasts. i) It measures the relative expression levels of OsMYB8 in the pistils of ZH11, OsMYB8TF/Osmyb8ZH transgenic lines, and Osmyb8ZH mutants. j) It displays the comparison of spikes between ZH11, OsMYB8TF/Osmyb8ZH transgenic lines, and Osmyb8ZH mutants at 11:30 AM in October 2022 in Guangzhou. k) It records the number of flowers opened at different time points in October for ZH11, OsMYB8TF/Osmyb8ZH transgenic lines, and Osmyb8ZH mutants.
Thus, the results highlight the role of natural variation in the genetic regulation of rice DFOT, especially how polymorphisms in the promoter region affect the expression of OsMYB8 and subsequent plant biological responses. These findings are crucial for understanding the timing regulatory mechanisms in rice growth and development, as well as providing important genetic information for rice breeding.
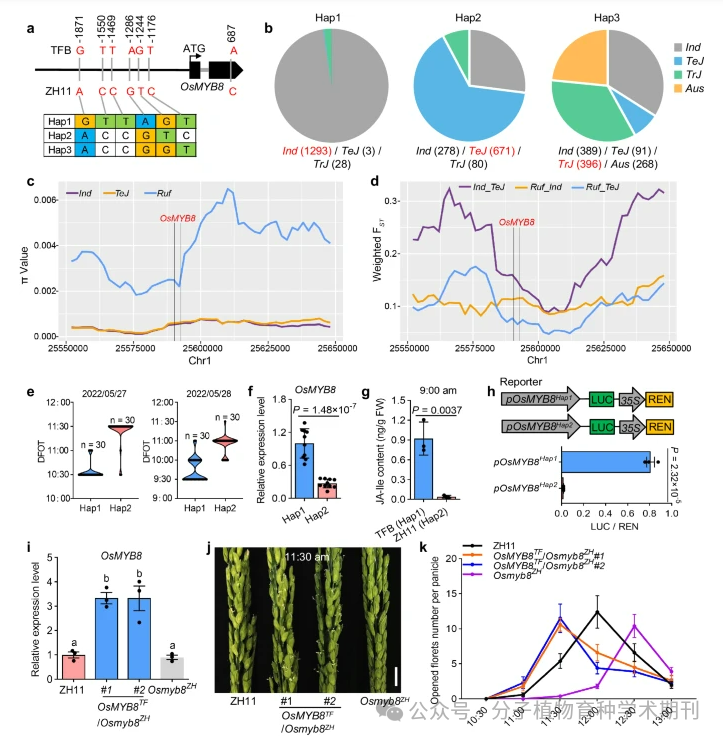
Figure 6 Natural variation in OsMYB8 promoter confers DFOT divergence in japonica and indica
Figure 6 Natural variation in OsMYB8 promoter confers DFOT divergence in japonica and indica
Figure 7 shows how the indica allele of the OsMYB8 gene promotes diurnal flower opening time (DFOT) in japonica rice. a) and d) compare the spikes of ZH11 with NIL^TFB and XS134 with CSSL9311 at 12:00 PM in Guangzhou, with the scale bar indicating 1 cm. b) and e) measure the number of flowers opened per spike for ZH11 and NIL^TFB, and XS134 and CSSL9311 at different daytime time points, with values representing mean and standard error (SEM) (n=10 spikes). c) and f) measure the relative expression levels of OsMYB8 and OsJAR1 in the pistils of ZH11 and NIL^TFB, and XS134 and CSSL9311, with values represented as mean ± standard error (SEM) (n=3 biological replicates). Significance is assessed using a two-tailed Student’s t-test and P values are indicated. g) A model depicting the OsMYB8 and OsJAR1 module illustrates how it regulates the DFOT differences between indica and japonica rice. The natural variation in the OsMYB8 promoter sequence leads to higher expression levels of OsMYB8 in indica, resulting in greater accumulation of JA-Ile, and consequently, earlier DFOT compared to japonica.
Through these results, Figure 7 clearly illustrates how differences in gene expression affect the genetic variation of flowering time in rice. These findings are crucial for understanding the crop biological clock and flowering mechanisms, and they provide potential pathways for improving crop productivity traits through gene editing.
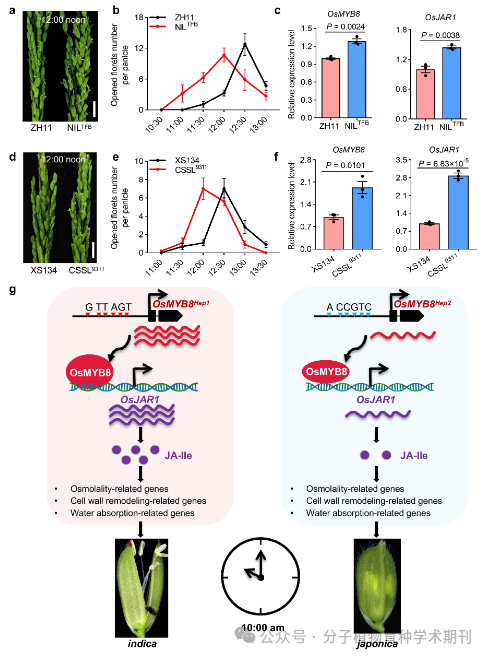
Figure 7 The indica allele of OsMYB8 promotes japonica DFOT
2 Analysis of Research Results
The research results show that through a comprehensive analysis of the interaction between OsMYB8 and OsJAR1 gene modules, it elucidates their roles in regulating rice cell permeability and cell wall remodeling, revealing how these two mechanisms lead to differences in flowering time in indica and japonica rice. The study further indicates that transferring the indica OsMYB8 allele into japonica rice can significantly advance the flowering time of japonica rice, which may provide an effective approach for improving the production of hybrid rice varieties. This finding not only deepens our understanding of the biology of plant flowering but also provides new molecular tools for rice breeding practices.
3 Research Evaluation
This study provides new insights into the genetic regulation of rice flowering time and offers feasible strategies for improving the breeding efficiency of hybrid rice through gene manipulation. This work not only enhances our understanding of the regulation of flowering time in plants but also provides new tools for the improvement of crops such as rice.
4 Conclusion
By deeply analyzing the interactions between OsMYB8 and OsJAR1, the molecular mechanisms regulating flowering time in rice are revealed. The application of gene editing technology successfully adjusted the flowering time of rice. This discovery has significant practical application value for enhancing the production efficiency of hybrid rice varieties.

Acknowledgments
Thanks to Nature magazine for providing readers with the valuable opportunity to access open access papers. This open access policy allows me to download, read, comment on, and disseminate this excellent research paper for free. Through such mechanisms, Nature magazine not only promotes the widespread dissemination of scientific knowledge but also provides valuable learning resources for researchers, students, and all individuals interested in science worldwide. This spirit of openness and sharing has immeasurable value in advancing scientific progress and fostering public understanding and interest in scientific research.
END

Scan to follow us!
Core journal of Chinese Science and Technology Papers Statistics Source
Core journal in the Overview of Core Journals of Peking University Library
Chinese Core Academic Journal (A) by the Chinese Science Evaluation Research Center (RCCSE)
Chinese Core Journal (A) by the Agricultural Information Research Institute of the Chinese Academy of Agricultural Sciences
Chinese Journal Fulltext Database (CJFD)
Chemical Abstracts (CA) by the American Chemical Society
Japan Science & Technology Corporation (JST) database
Address: Room 13B, Shuangdao Apartment, 128 Haixiu Avenue, Haikou City, Hainan Province
Postal Code: 570206
Phone: 0898-68966415
Website: http://molplantbreed.org
Email: [email protected]
Click here to read the original article