We received a question from a bioinformatics beginner marathon student, what should we do!!! Of course, we should pamper her! The question is as follows:

First, I searched for relevant information
Using keywords to search in WeChat: MSigDB database glycolysis-related genes. I found an article published in October 2022 in the journal Frontiers in Endocrinology: Identification of risk model based on glycolysis-related genes in the metastasis of osteosarcoma. This article used a collection of glycolysis-related pathway genes:
We obtained 5 glycolysis-related pathway gene sets from MSigDB, namely:
BIOCARTA GLYCOLYSIS PATHWAY GO GLYCOLYTIC PROCESS, HALLMARK GLYCOLYSIS, KEGG GLYCOLYSIS GLUCONEOGENESIS, REACTOME GLYCOLYSIS.
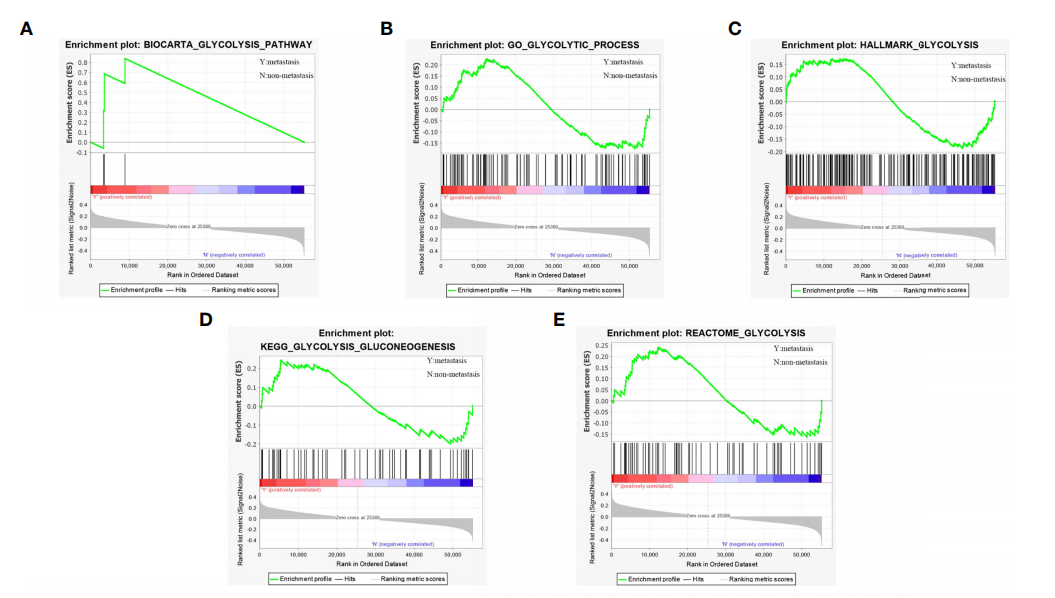
I feel there should be more than 5 gene sets!
I also saw another article published in August 2023 in the journal Cancer Cell Int titled Glycolysis-related biomarker TCIRG1 participates in regulation of renal cell carcinoma progression and tumor immune microenvironment by affecting aerobic glycolysis and AKT/mTOR signaling pathway, which also provided specific instructions on how to search for glycolysis-related genes from the MSigDB database:

Of course, we can directly download the supplementary table table2 from this article, but the MSigDB database underwent a major update in 2024, as mentioned in the article: What are the reasons for the failure to obtain gene sets from the msigdbr database?, we still recommend checking the latest data!

The article found 21 pathways:
| BIOCARTA_ETC_PATHWAY |
|---|
| BIOCARTA_FEEDER_PATHWAY |
| BIOCARTA_GLYCOLYSIS_PATHWAY |
| BIOCARTA_KREB_PATHWAY |
| CHEN_LUNG_CANCER_SURVIVAL |
| DCA_UP.V1_DN |
| DCA_UP.V1_UP |
| GOBP_FRUCTOSE_1_6_BISPHOSPHATE_METABOLIC_PROCESS |
| GOBP_LACTATE_TRANSMEMBRANE_TRANSPORT |
| GOMF_LACTATE_TRANSMEMBRANE_TRANSPORTER_ACTIVITY |
| HALLMARK_GLYCOLYSIS |
| KEGG_CITRATE_CYCLE_TCA_CYCLE |
| KEGG_GLYCINE_SERINE_AND_THREONINE_METABOLISM |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS |
| MODULE_306 |
| REACTOME_GLYCOLYSIS |
| REACTOME_REGULATION_OF_GLYCOLYSIS_BY_FRUCTOSE_2_6_BISPHOSPHATE_METABOLISM |
| WP_AEROBIC_GLYCOLYSIS |
| WP_GLYCOLYSIS_AND_GLUCONEOGENESIS |
| WP_GLYCOLYSIS_IN_SENESCENCE |
| WP_HIF1A_AND_PPARG_REGULATION_OF_GLYCOLYSIS |
Let’s see how to implement it with code
We will use the keyword glycolysis to search the MSigDB database: https://www.gsea-msigdb.org/gsea/msigdb/index.jsp.
First, download the entire library, the file is less than 30M:

library(clusterProfiler)
library(org.Hs.eg.db)
library(GSEABase)
## === All pathways
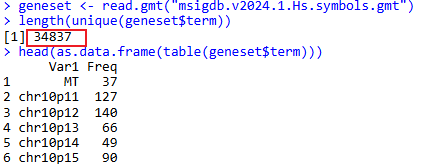
geneset <- read.gmt("msigdb.v2024.1.Hs.symbols.gmt")
length(unique(geneset$term))
head(as.data.frame(table(geneset$term)))
There are a total of 34837 gene sets:
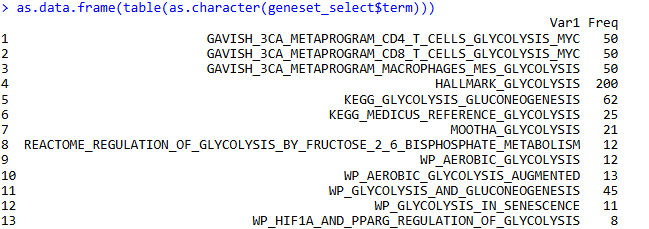
Only found 13 pathways, which is fewer than the above article. Looking at the pathways above, some gene sets do not contain the keyword glycolysis:
# Search for glycolysis-related gene sets: glycolysis
geneset_select <- geneset[grep(pattern ="glycolysis", geneset$term,ignore.case = T),]
str(geneset_select)
as.data.frame(table(as.character(geneset_select$term)))
Web Version Search and Download
The above code can only retrieve gene sets with the keyword in their names; there may be other gene sets that do not have this keyword but have glycolysis-related functions. See the method below to obtain the file: genesets.v2024.1.Hs.gmt, which has 22 gene sets, one more than the article above:

Read it in R to see:
### Web Download
geneset <- read.gmt("genesets.v2024.1.Hs.gmt")
length(unique(geneset$term))
as.data.frame(table(geneset$term))

This way, we have obtained all the glycolysis-related genes, and we can happily use them later!
If you have similar questions, you can check the links below:
Bioinformatics Introduction & Data Mining Online Live Course January 2025
After 5 years, our Bioinformatics Skills Tree VIP apprentices are continuing to recruit!
A low-cost solution to meet your bioinformatics analysis computing needs.