Click the blue text to follow us
Computer-Aided Diagnosis System Based on Deep Learning
Design and Implementation in Embedded Systems
Design and implementation
of computer aided diagnosis
system based on deep learning
in embedded system
Author Affiliations
Zhang Lili, Yang Jinzhu, Shan Yufu,
Zhang Gaoyuan, Xin Junchang
Northeastern University, College of Computer Science and Engineering
National Computer Experimental Teaching Demonstration Center,
Shenyang, Liaoning 110819, China
ZHANG Lili, YANG Jinzhu, SHAN Yufu,
ZHANG Gaoyuan, XIN Junchang
National Computer Experimental Teaching
Demonstration Center,
College of Computer Science and
Engineering, Northeastern University,
Shenyang 110819, China
Author Biography:
Zhang Lili (1984—), female, from Fuxin, Liaoning, senior experimentalist, main research directions include communication technology, image processing, experimental technology management, etc.Corresponding Author: Yang Jinzhu (1979—), male, from Tongliao, Inner Mongolia, professor, main research directions include artificial intelligence, image processing analysis, etc.
Below is the structure of this article
Click the image to view the larger version
Abstract
With the improvement of modern medical imaging quality and the increase of medical image data, it takes more and more time and cost for doctors to quickly find the designated category medical image data from massive medical images. Computer-aided diagnosis of medical image data in the medical image database has become a very realistic demand. Therefore, as a typical case of artificial intelligence experimental application, this paper designs a computer-aided diagnosis system that can detect the location of lesion area by using improved deep learning algorithm. The system can complete the automatic segmentation of various organs in medical images. A convolution neural network framework is established and used to train a large number of data in medical image database. Then, according to the trained classifier model, the images are classified into corresponding categories, thus completing the intelligent diagnosis process. Finally, the system is transplanted to the embedded development board, which is convenient for the subsequent development of actual products. The experimental results show that the system can better identify pathological areas, can be applied to computer-aided diagnosis of X-ray chest film, and provide better diagnostic help for radiologists.
Keywords: deep learning; computer
aided; medical imaging; convolution
neural network; embedded
Below is the full text for reading
In recent years, malignant tumors have become the biggest killer threatening the safety of human life globally. In developed countries, about 70% of cases diagnosed as lung cancer are usually at an advanced stage, making early detection and treatment of lung cancer very important. Due to the increasing size of lung nodules that can evolve into cancer cells, the size of lung nodules is the main basis for lung cancer diagnosis. Medical imaging refers to the technology and processing process of obtaining internal tissue images of the human body or parts of the human body in a non-invasive manner for medical or medical research. In the field of medical imaging, the examination methods for lung nodules include X-ray examination, CT examination, radioactive nuclide examination, MR examination, etc. With the advancement of computer technology and continuous breakthroughs in medical imaging technology, medical image analysis and diagnosis have entered the era of big data. Medical image-based computer-aided diagnosis systems (MIBCAD) can quickly assist doctors in diagnosing patients’ conditions by collecting a large amount of medical image data information from a certain type of patients, utilizing confirmed case information, current clinical diagnostic experience of doctors, and current patient medical history information, while reducing the influence of subjective factors.
Computer-aided medical image diagnosis involves three steps: collection, processing, and analysis to obtain the final diagnosis result. The main work of the collection step is to gather and organize patient data; processing refers to the handling of medical information, such as quantifying medical images; analysis represents statistical analysis and integration.
However, due to the lack of standards for medical image feature extraction and the difficulty of extraction, as well as the generally low contrast of clinical medical images and the unclear lesion areas, this poses significant challenges for the research of fully automated medical image classification technology. In recent years, deep learning has been applied to image classification problems, allowing meaningful feature representations to be learned from a large number of input images through specific network structures and training methods for subsequent classification or other visual tasks, one important application being the use of convolutional neural networks for image classification. As the amount of machine-readable clinical data in the field of medical imaging continues to grow, modern neural networks have shown exceptional performance in starting medical image classification tasks. Literature [3] suggests that modern neural networks have unique advantages in solving problems related to natural images and videos. Literature [4] also indicates that modern neural networks are rapidly becoming the standard for medical image classification, detection, and segmentation tasks for input forms such as CT, MRI, X-ray, and ultrasound. Literature [5] proposes that improved neural networks can address the challenges posed by medical imaging and optimize the processing of medical images and related issues after reviewing current research on computer-aided medical diagnosis, edge detection, and medical image segmentation. Literature [6] further provides evidence that neural network-based models lead in most medical imaging diagnosis challenges. The majority of successful applications of neural networks in medical imaging diagnosis heavily rely on convolutional neural networks, first proposed by Cun et al. For anomaly detection and segmentation, the most popular variants are UNets and VNets, both based on the concept of fully convolutional networks introduced in [8]. Representative examples of neural network model applications from medical literature in medical image classification and assisted diagnosis include: skin cancer classification [9], diabetic retinopathy [10], X-ray tuberculosis detection [11], and chest CT diagnosis of lung cancer [12]. After extensive data collection and literature review, this paper systematically implements the project focusing on dataset acquisition, selection of deep learning methods, establishment of deep learning environments, and specific practical applications.
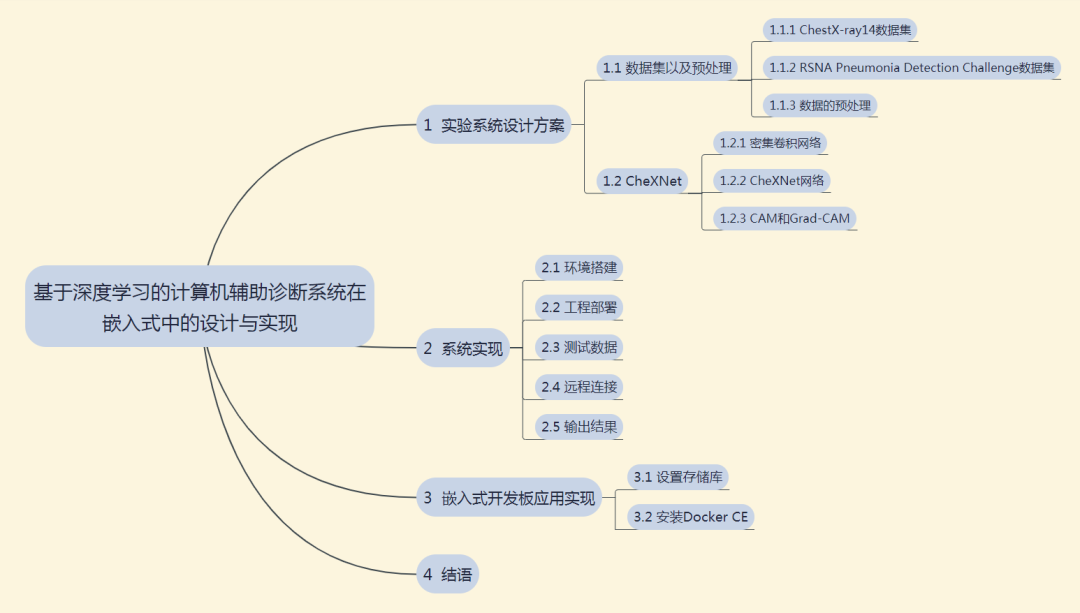
1 Experimental System Design Scheme
This section mainly introduces the dataset and preprocessing, and the CheXNet network.
1.1 Dataset and Preprocessing
In this project, two datasets were mainly used, namely the ChestX-ray14 dataset and the latest RSNA Pneumonia Detection Challenge dataset released by Kaggle [13-14]. Below is a brief introduction to these two datasets.
1.1.1 ChestX-ray14 Dataset
The ChestX-ray dataset was collected, annotated, and released by the National Institutes of Health (NIH) in the United States. This dataset contains 112,120 frontal view X-ray images from 30,805 patients, along with image labels for 14 diseases mined from relevant radiology reports using NLP (each image can have multiple labels). The dataset includes 14 common chest pathologies, including atelectasis, consolidation, infiltration, pneumothorax, edema, emphysema, fibrosis, effusion, pneumonia, pleural thickening, cardiomegaly, nodules, masses, and hernias.
1.1.2 RSNA Pneumonia Detection Challenge Dataset
In early September, the Radiological Society of North America (RSNA) launched a pneumonia detection competition in collaboration with Kaggle. In the first phase, the competition has released a dataset of nearly 30,000 frontal chest X-ray images. These images were also selected and annotated from the publicly available lung medical image dataset at the NIH Clinical Center (NIHCC).
Unlike the previous dataset, this dataset does not specify the type of lesions but only indicates whether pneumonia is present. For approximately 9,000 images with pneumonia, the dataset provides the coordinates of the lower left corner and the width and height of the lesion area annotated by radiologists. Participants need to determine whether pneumonia is present in the test set (1,000 images) and provide the coordinates and range of the lesion area.
1.1.3 Data Preprocessing
Both datasets mentioned above are written in DICOM protocol, with the image format being DCM. For training deep learning models, DCM format images cannot be used directly and must be converted. Traditionally, C or MATLAB programming is used to convert DCM format to a directly openable format, which involves a large amount of code and is inefficient. However, Python includes the pydicom package, which can directly read the pixel data of DCM format images, and with the help of OpenCV, batch conversion to JPG format can be performed. The converted images are shown in Figure 1.
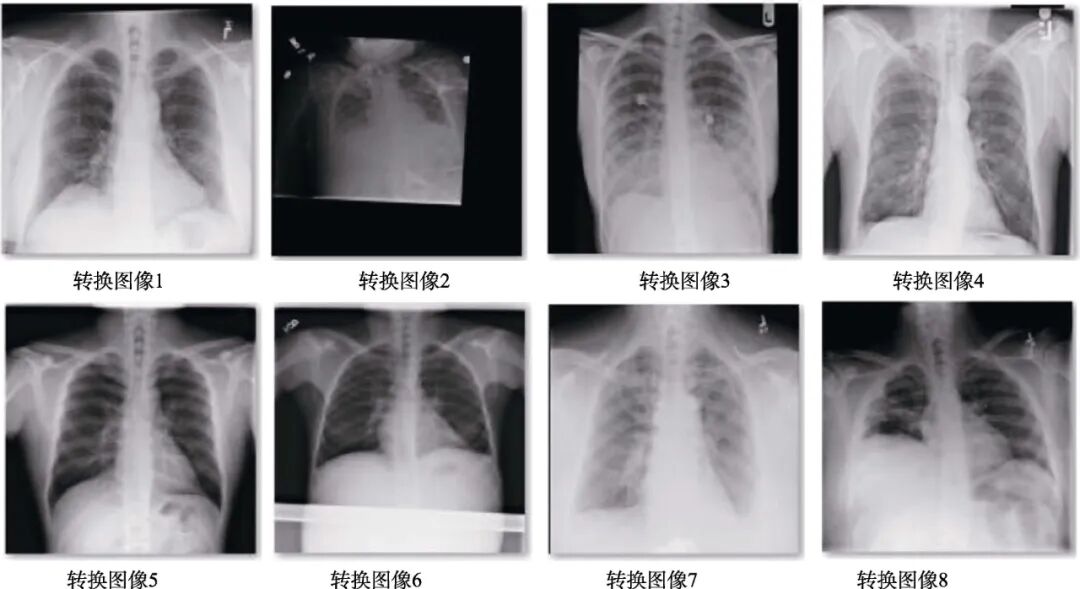
Figure 1 Images after data format conversion
1.2 CheXNet
1.2.1 Dense Convolutional Network
The Dense Convolutional Network (DenseNet) is one of the latest CNN structures, which adopts a novel connection mode based on cross-layer connections on top of traditional convolutional neural networks. This connection mode links the front and back layers of the network, allowing signals to flow rapidly between the input and output layers [15]. In a densely connected convolutional network, all layers are connected pairwise, with each layer receiving additional inputs from all previous layers and passing its own feature maps to all subsequent layers. This ensures maximum information flow between layers in the network, allowing the feature space of input data to be reused repeatedly.
This paper uses a 121-layer Dense Convolutional Network (DenseNet-121) to train the ChestX-ray14 dataset. DenseNet-121 has 4 dense blocks, each with the same number of layers, where each “conv” layer corresponds to the sequence BN-relu-Conv. Before entering the first dense block, a convolution with 64 output channels is performed on the input image. For convolution layers with a kernel size of 3×3, one pixel is padded on each side of the input to maintain the feature map size. Then, a 1×1 convolution layer and a 2×2 average pooling layer are used as transition layers between two consecutive dense blocks. At the end of the last dense block, global average pooling is performed, followed by a softmax classifier.
The network configuration for X-ray chest film auxiliary diagnosis based on dense convolution involves training a 121-layer Dense Convolutional Network (DenseNet-121) on the ChestX-ray14 dataset, achieving similar results to the Chex121 network, which includes 4 dense blocks and 3 transition layers. The network architecture can be viewed under the GRAPH section of TensorBoard, with the dense blocks shown in Figure 2 and the transition layers in Figure 3. Due to the depth of the network, only one dense block and one transition layer structure are provided here.
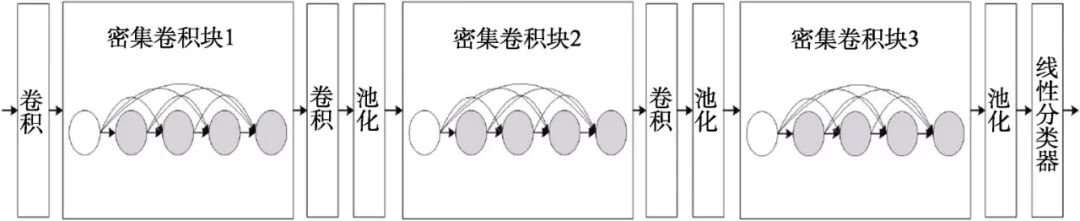
Figure 2 Deep DenseNet with 3 dense blocks
Figure 3 DenseNet used for training
1.2.2 CheXNet Network
Currently, chest X-ray examination is the best method for diagnosing pneumonia, playing a crucial role in clinical care and epidemiological research. However, detecting pneumonia through X-ray images is a challenging task that relies on the expertise of radiologists. The CheXNet algorithm proposed by Andrew Ng’s team is a 121-layer convolutional neural network capable of determining whether a patient has pneumonia from chest X-ray images, surpassing the performance of professional radiologists [16].
The CheXNet model, as shown in Figure 4, is a 121-layer convolutional neural network. The input of the model is chest X-ray images, and the output is the probability of pneumonia and a heatmap to locate the image areas most indicative of pneumonia. The CheXNet was trained using the recently released ChestX-ray14 dataset, which contains 112,120 individually annotated frontal chest X-ray images of 14 different chest diseases (including pneumonia). This deep network is optimized using dense connections and batch normalization.
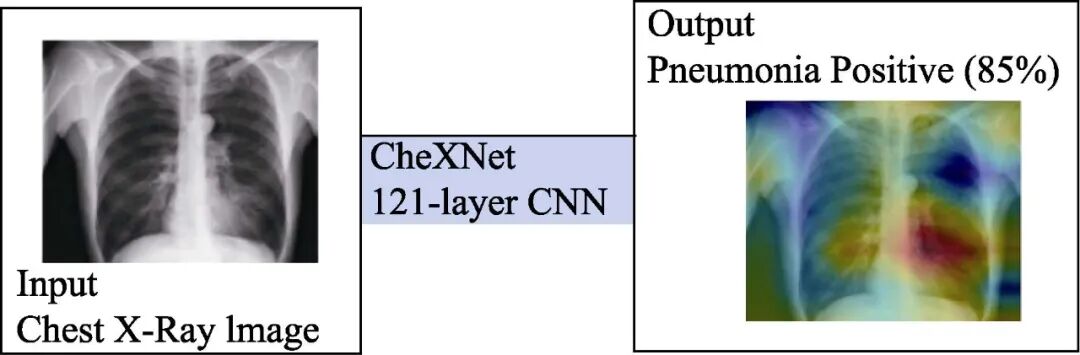
Figure 4 CheXNet Model
1.2.3 CAM and Grad-CAM
CAM (class activation mapping) is used to locate key parts of images, while Grad-CAM uses the gradients of the last convolutional layer to generate heatmaps that highlight important pixels in the input image for classification. CAM replaces the last fully connected layer of the network with GAP (global average pooling), then sets the output channels of the last convolutional layer to match the number of categories to be classified. This way, the trained network has a one-dimensional vector representation of weights for each category corresponding to the number of convolution output channels, allowing for the weighted accumulation to obtain interpretable regions.
2 System Implementation
2.1 Environment Setup
This project configures a deep learning-based medical image auxiliary diagnosis system on a GPU server.
During installation, Python 3.6 is required, and pip must also be installed according to the version corresponding to the default Python. In the actual configuration process, some dependencies may not work properly, and after upgrading to the latest versions, compilation was successful. The actual usage was through the Python 3.6.6 environment generated by Anaconda, with pip installing the above items. Based on error messages, Keras==2.1.6, tensorflow-gpu==1.4.0, opencv-python==3.4.3 were updated, thus meeting the environment requirements.
2.2 Project Deployment
The project references the CheXNet-Keras contributed by brucechou1983. The entire project was downloaded using git, and relevant folders were established according to the config.ini provided by the author. After downloading the pre-trained model to the specified location, deployment was completed. The file structure is shown in Figure 5.
Figure 5 File Structure Display
2.3 Test Data
(1) First, create a test.csv for the data to be tested, then copy the corresponding images into data/images.
(2) Execute python cam.py, the command line displays the entire function call and execution process, including initialization, loading model parameters, sequentially reading the images to be tested, performing cam processing, and outputting results.
(3) The entire process takes about 26 seconds. About 10 seconds are spent loading the model and reading the test data. After reading is complete, processing 22 images takes 16 seconds, averaging 0.7 seconds per image, with a speed close to 1 frame/s.
2.4 Remote Connection
Using deep learning methods for medical image processing, if running on a CPU, the time required to recognize and segment an image may take more than ten seconds. For a DenseNet such as this, loading the model and classification process will take even longer. However, when utilizing GPU acceleration, the time to process an image is greatly reduced. Ordinary laptops have their own limitations, such as low voltage CPUs, entry-level graphics cards, or no graphics card, so this experiment uses remote login to access the GPU server, completing deep learning tasks using the configured GPU server.
TeamViewer is used for remote control of the GPU server. To connect to another computer, both computers need to run TeamViewer simultaneously. The software automatically generates partner IDs on both computers the first time it starts, and you just need to enter the partner ID into TeamViewer to establish a connection immediately. TeamViewer’s remote connection is supported by the TeamViewer server relay, so it is not limited by network segments. Tests have shown that a laptop connected via a mobile hotspot can control a GPU server on a campus network, but there is some latency.
2.5 Output Results
First, the images with labels are output, meaning that classification is performed simultaneously with segmentation. The test results are displayed in heatmap form, with some sample pathological occurrence area recognition results shown in Figure 6. Areas with confidence below 0.2 are ignored, and the remaining parts indicate areas with inflammation or potential inflammation. The darker the color, the higher the probability of lesions in that area. For the DenseNet121 used by CheXNet, when inputting a 1024×1024 image, the image size is reduced to 224×224 before being fed into the neural network. In subsequent work, the output image is restored to the input size through upsampling to accurately determine the specific location of the lesion area. As can be seen from the figure, although there are some deviations in the recognition of pathological areas, the network can already identify pathological areas well, which can be applied to computer-aided diagnosis of X-ray chest films, providing better diagnostic assistance for radiologists.
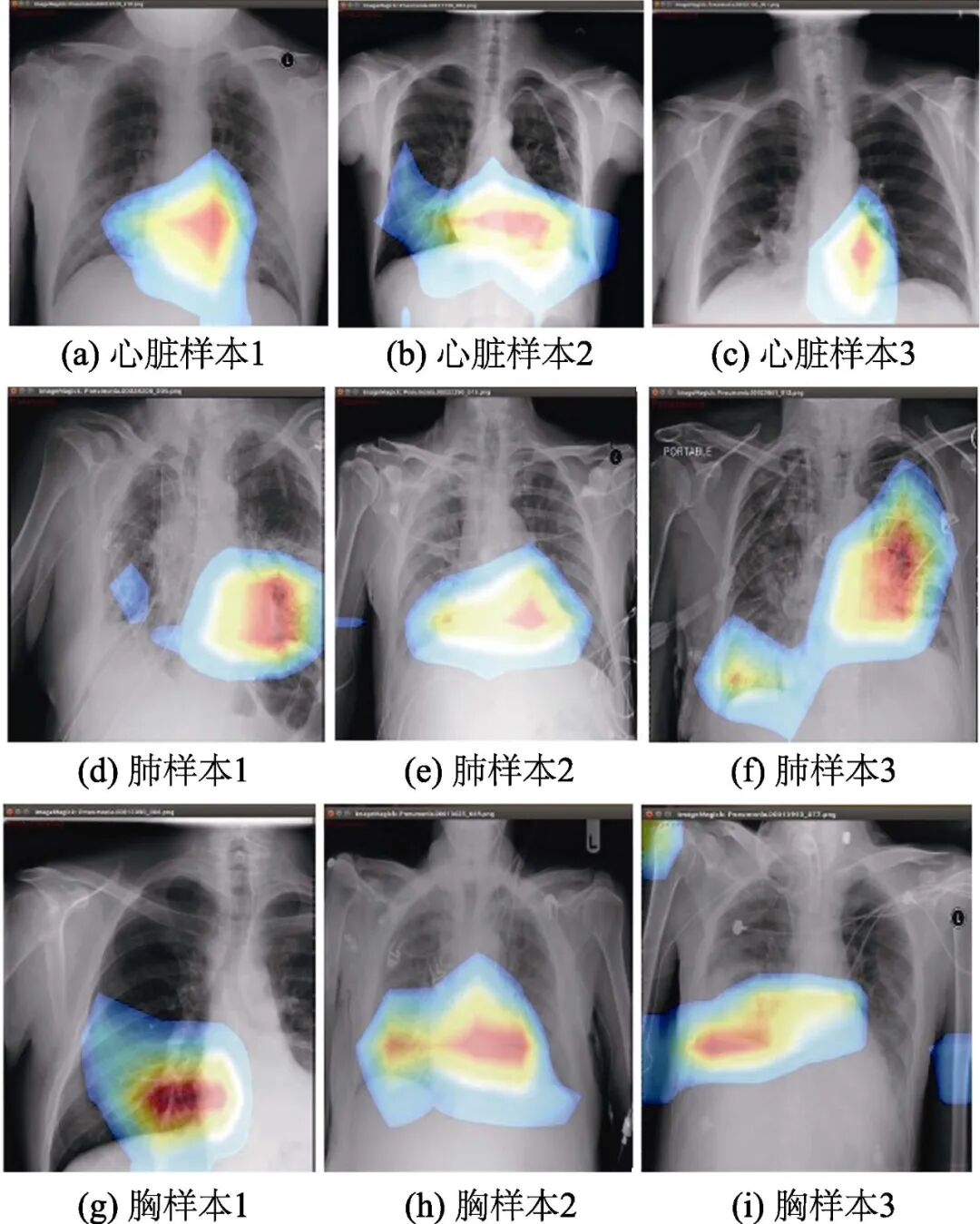
Figure 6 Recognition results of some samples
3 Implementation of Embedded Development Board Application
After completing the above PC-based medical image auxiliary diagnosis, the above functions are implemented on the embedded development board. Since the Nvidia Jetson tk1 embedded development board uses an ARM architecture CPU, the operating system must be compiled on an Ubuntu 14.04 host and then written to the storage. To deploy the system to the tk1 embedded development board, it is necessary to flash the machine, install Ubuntu, and configure CUDA and CUDNN. Finally, configure Docker to run CheXNet using the mounted image.
3.1 Setting Up the Repository
1) Update the apt package index.
$ sudo apt-get update.
2) Install packages to allow apt to use repositories over HTTPS.
$ sudo apt-get install \
apt-transport-https \
ca-certificates \
curl \
software-properties-common.
3) Add Docker’s official GPG key.
$ curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add – . Verify that you now have the key with the fingerprint by searching for the last 8 characters of the fingerprint.
4) Set up the stable repository using the following command.
Even if you want to install builds from the edge or test repository, you always need the stable repository.
$ sudo add-apt-repository “deb [arch=armhf] https://download.docker.com/linux/ubuntu \
$(lsb_release -cs) \
stable”.
3.2 Installing Docker CE
1) Update the apt package index.
$ sudo apt-get update.
2) Install the latest version of Docker CE, or go to the next step to install a specific version.
$ sudo apt-get install docker-ce.
3) To install a specific version of Docker CE, list the available versions in the repo, then select and install.
List the available versions in the repo:
$ apt-cache madison docker-ce
docker-ce|18.03.0~ce-0~ubuntu|
https://download.docker.com/linux/ubuntu
xenial/stable amd64 Packages.
Install a specific version by its fully qualified package name (docker-ce) and the “=” version string (second column), for example, docker-ce=18.03.0~ce-0~ubuntu.
$ sudo apt-get install docker-ce=
4) Verify that Docker CE is installed correctly by running the hello-world image.
Run the command “$ sudo docker run hello-world” to download a test image and run it in a container. When the container runs, it will print a message and exit.
For promotional applications, the CheXNet project and its required environment, dependencies, etc., are mounted in the Docker image. Any computer with a Linux operating system and Nvidia graphics hardware drivers can directly deploy this image after configuring Docker, greatly reducing the workload of deploying deep learning environments.
4 Conclusion
With the development of computer technology and artificial intelligence, a large number of automatic classification technologies have emerged, enabling computer-aided medical image diagnosis. This paper designs and implements a deep learning-based computer-aided diagnosis system based on existing research, achieving medical image classification, detection, and segmentation through the configuration of a caffeTensorFlo-based development and testing environment, and transplanting it to an embedded development board. The system can significantly enhance the efficiency of doctors’ diagnoses while reducing the influence of subjective factors, playing a significant role in improving diagnostic accuracy, reducing missed diagnoses, and enhancing work efficiency, which has special significance for clinical medicine.
References
[1] KUO W J. Computer-aided diagnosis for feature selection and classification of liver tumors in computed tomography images[C]// 2018 IEEE International Conference on Applied System Invention (ICASI). Chiba, Japan: IEEE, 2018, 1207–1210.
[2] CHEN X Q, CAO C J, MAI J B. Network anomaly detection based on deep support vector data description[C]// 2020 5th IEEE International Conference on Big Data Analytics (ICBDA). Xiamen, China: IEEE, 2020, 251–255.
[3] Huang Jiangshan, Wang Xiuhong. A review of the application of deep learning in medical image analysis[J]. Library and Information Research, 2019, 12(2): 92–98.
[4] WANG Y, GE X K, Ma H, et al. Deep learning in medical ultrasound image analysis: A review[J]. IEEE Access, 2021, 9: 54310–54324.
[5] JIANG J, TRUNDLE P, REN J. Medical image analysis with artificial neural networks[J]. Computerized Medical Imaging and Graphics, 2010, 34(8): 617–631.
[6] Zhou Tao, Huo Bingqiang, Lu Huiling, et al. Research progress on residual neural network optimization algorithms for medical image disease diagnosis[J]. Chinese Journal of Image and Graphics, 2020, 25(10): 2079–2092.
[7] CUN Y L, BOTTOU L, BENGIO Y. Gradient-based learning applied to document recognition[J]. Proceedings of the IEEE, 1998, 86(11): 2278–2324.
[8] LONG J, SHELHAMER E, DARRELL T. Fully convolutional networks for semantic segmentation[C]// 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Boston, MA, USA: IEEE, 2015, 3431–3440.
[9] ESTEVA A, KUPREL B, NOVOA R A. Dermatologist-level classification of skin cancer with deep neural networks[J]. Nature, 2017, 542(7639): 115–118.
[10] GULSHAN V, PENG L, CORAM M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs[J]. Jama, 2016, 316(22): 2402–2410.
[11] KARHAN Z, AKAL F. Covid-19 Classification using deep learning in chest X-Ray images[C]// 2020 Medical Technologies Congress (TIPTEKNO). Antalya, Turkey: IEEE, 2020, 1–4.
[12] Xu Jianguo. Discussion on the clinical effect and detection rate of low-dose chest CT scans for lung cancer diagnosis[J]. China Continuing Medical Education, 2021, 13(23): 147–149.
[13] HO T K K, GWAK J H. Utilizing knowledge distillation in deep learning for classification of chest X-Ray abnormalities[J]. IEEE Access, 2020, 8, 160749–160761.
[14] WANG X S, PENG Y F, LU L, et al. ChestX-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases[C]// 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, HI, USA: IEEE, 2017, 3462–3471.
[15] Song Jiafei, Song Xinxia, Yang Hequn, et al. Fully convolutional pooling algorithm based on dense convolutional neural networks[J]. Intelligent Computer and Applications, 2021, 11(3): 66–69.
[16] RAJPURKAR P, IRVIN J, ZHU K, et al. Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning[Z]. arXiv, preprint arXiv: 1711.05225v3, 2017.
Cite this article: ZHANG L L, YANG J Z, SHAN Y F, et al. Design and implementation of computer aided diagnosis system based on deep learning in embedded system[J]. Experimental Technology and Management, 2022, 39(8): 49-54. (in Chinese)
Experimental Technology and Management, 2022, Issue 8, P49-54

Experimental Technology and Management
Comprehensive Science and Technology Journal · Journal of Laboratory Work Research Association of Higher Education Institutions
Chinese Core Journal · RCCSE Chinese Authoritative Academic Journal (A+)
Excellent Science and Technology Journal of Chinese Universities · First Excellent Journal Implementing “CAJ-CD Standard”
Journal Website · Online Submission
http://syjl.cbpt.cnki.net
http://syjl.chinajournal.net.cn
Contact & Subscription
Editorial Office Phone: 010-62783005 or 010-62797828
Email: [email protected]
Subscription and Distribution Phone: 010-62792635
Email: [email protected]
Click
To download the full text, please click Read the original text