
Live cell in vitro functional studies play a crucial role in basic biology, drug mechanism research, and disease control model establishment. The microenvironment in which cells reside affects their health status and phenotype. Therefore, breaking through the limitations of traditional static and large-space live cell culture methods under in vitro conditions by establishing a more precise dynamic control system (temperature, gas, liquid flow) in a closed space undoubtedly elevates live cell functional research and overall cell biology research to a new level. The CellASIC® ONIX2 is specifically designed for this gap, significantly surpassing the limitations of traditional methods, highly recreating the in vivo microenvironment, and perfectly combining cell culture with functional analysis to achieve a unique experimental approach.
Components of the Microfluidic Platform

Illustration of CellASIC® ONIX2 paired with a microscope
Closed Microenvironment Allows Flexible Experimental Conditions
Different cell types have different requirements for their growth environment. The CellASIC® ONIX2 microfluidic culture plates are uniquely designed for mammals, bacteria, yeast, and more, optimizing the cell growth environment. In live cell dynamic monitoring, especially in long-term continuous observation experiments, it ensures a stable and favorable microenvironment for cell growth, meeting your various experimental needs.
Application Directions

Experimental Cases
The CellASIC® ONIX2 has a flexible and wide range of applications, with chips designed for different samples, divided into bacterial chips, mammalian cell chips, and yeast chips. These chips are suitable for various sample types, such as: primary cells, tumor cell lines, stem cell lines, adherent/suspended cells, small tissues/cell clusters, and extracellular matrix.
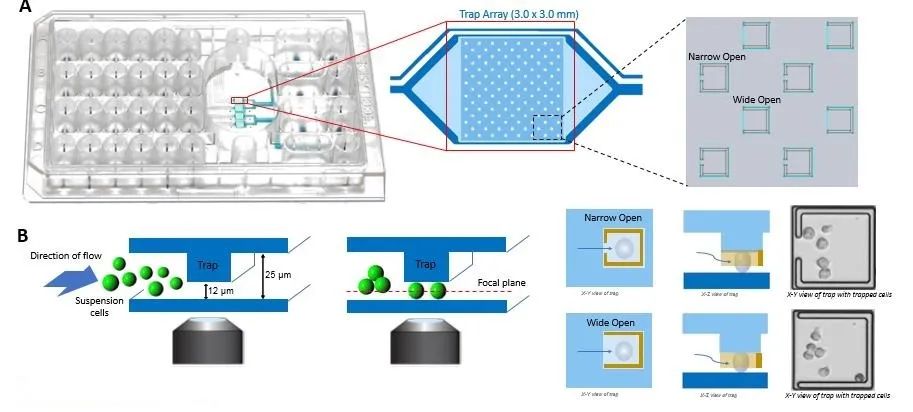
Suspension Cell Culture
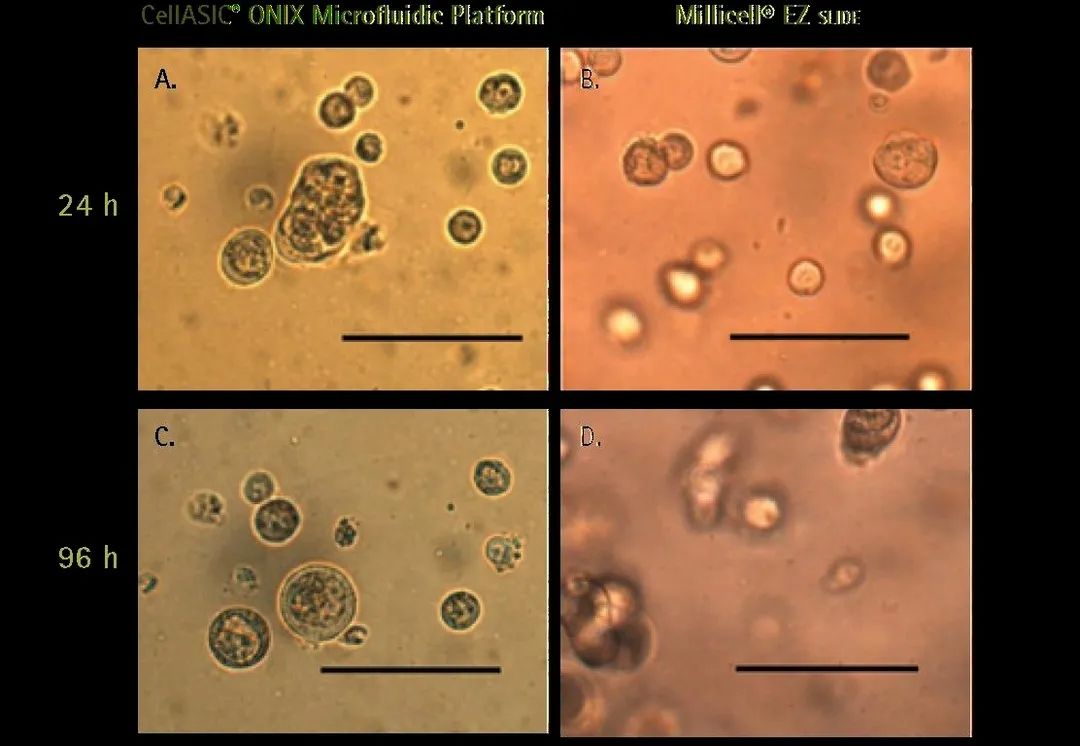
3D Cell Culture
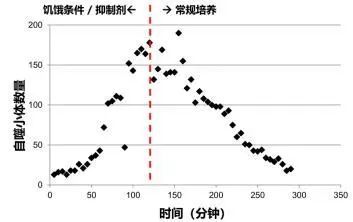
Cell Autophagy Quantitative Analysis Detection:
1. Cell starvation + lysosome inhibition (formation of autophagosomes)
2. Recovery process after adding complete culture medium (lysosome degradation)
3. Data analysis through image acquisition
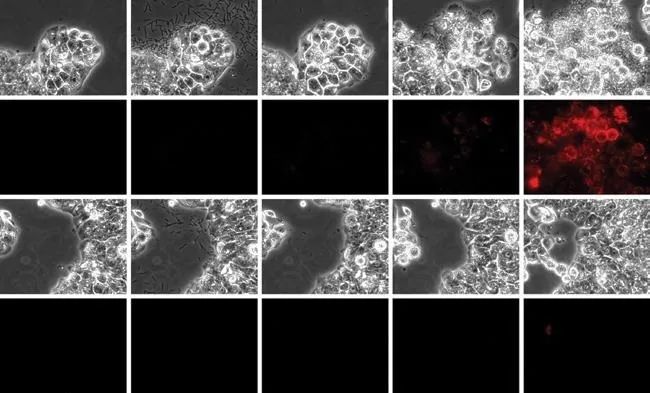
Host-Pathogen Interaction:
1. Precise perfusion control
2. Clear imaging time
3. Differential delivery of infectious pathogens and drugs
4. Long-term culture
5. Real-time imaging
References
1. Burke T. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Current Biology, March 2014; 24 (5): 579-85.
2. Sieger B. The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Molecular Microbiology, December 2013; 90 (5): 966-82.
3. Gordon AJ. Heritable change caused by Transient Transcription. PLoS Genetics. 2013 Jun; 9 (6): e1003595
4. Meyer R. Mps1 and lpl1/Aurora B Act Sequentially to Correctly Orient Chromosomes on the Meiotic Spindle of Budding Yeast. Science. 2013 March; 339, 1071.
5. Young J. Rate of environmental change determines stress response specificity. PNAS, 2013 March; 110 (10): 4140-4145.
6. Donovan C. Chromosome segregation impacts on cell growth and division site selection in Corynebacterium glutamicum. PLOS One, February 2013; 8(2): eSS078.
7. Rafelski SM. Mitochondrial network size scaling in budding yeast. Science, 2012 November; 338 (6108): 822-824.
8. Ludington W. Organelle size equalization by constitutive process. Current Biology, November 2012; 22 (22): 2173-2179.
9. Kraft C. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg regulates autophagy. EMBO J., 2012 Sept 12; 31 (18): 3691-703.
10. Sanchez-Diaz A. The Mitotic Exit Network and Cdc14 phosphatase initiate cytokinesis by counteracting CDK phosphorylations and blocking polarized growth. EMBO J., 2012 Aug 29; 31 (17): 3620-34.