
Introduction
Hello everyone, the literature I am sharing today is an article published on December 26, 2024, in ACS Nano: “A DNA Origami Pivot Hinge Driven by DNA Intercalators”. The corresponding author of this paper is Professor Do-Nyun Kim from Seoul National University, South Korea.
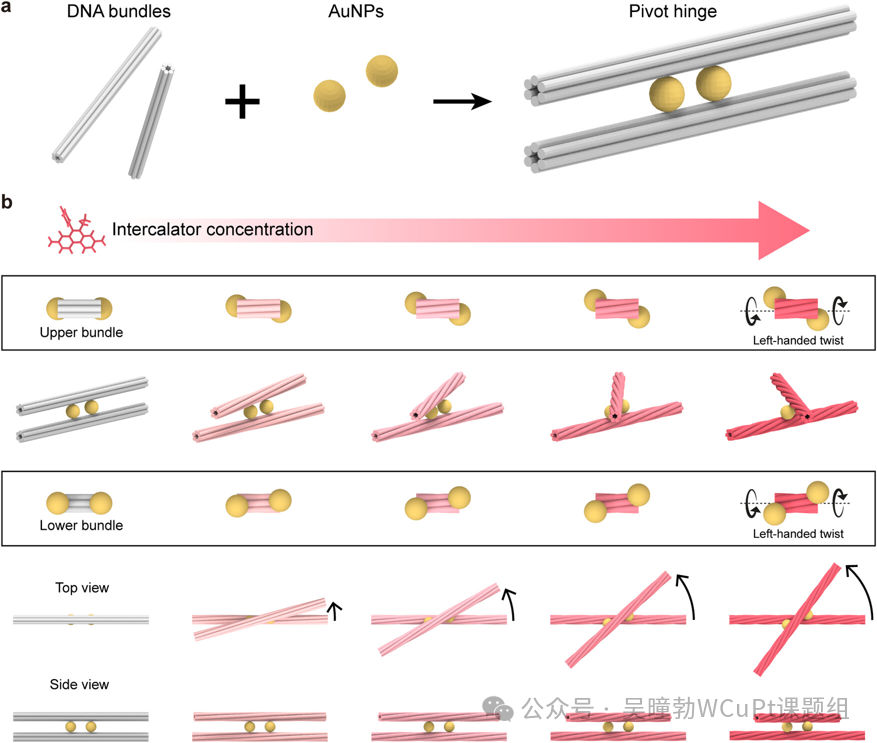
This study proposes a DNA origami hinge mechanism driven by DNA intercalators, achieving continuous and reversible rotational motion. The mechanism connects two six-helix bundles (6HB) through two gold nanoparticles (AuNPs), utilizing the twisting deformation induced by DNA intercalators to convert it into the rotational motion of the hinge. By varying the type and concentration of the intercalators, as well as the distance between the AuNPs, precise control over the sensitivity and range of hinge motion is achieved. Furthermore, this study demonstrates the potential of using this hinge to construct curved nanoactuators. This mechanism provides new ideas for the development of nanosensors and actuators, promising to advance the application of nanotechnology in fields such as biomedicine and materials science.
Scientific Issues
To achieve dynamic reconfiguration of nanostructures, existing studies have employed various methods, such as dynamic changes based on DNA strand displacement, and environmental stimuli (light, pH, temperature, etc.) triggering the reconfiguration of DNA origami nanostructures. However, these methods are often limited to achieving dynamic changes between a few (e.g., open and closed) configurational states, which presents certain limitations.
This study aims to achieve continuous dynamic changes in DNA origami structures through intercalator-induced DNA twisting deformation, overcoming the limitations of traditional methods that are confined to a limited number of states, providing a multifunctional approach for the development of nanoscale machines and robots.
Evaluation
Scientific Issue Dimension
The authors point out that traditional techniques based on DNA hybridization have limitations in dynamic reconfiguration, only allowing changes between a few configurational states. This study achieves continuous dynamic changes in DNA origami structures through intercalator-induced DNA twisting deformation, providing a new approach to address this issue. Although previous studies have utilized intercalators to regulate the twisting of DNA bundles to achieve dynamic changes in structures, applying this to construct DNA origami hinges with continuous rotational motion and further extending it to the construction of nanoactuators remains innovative and significant.However, this paper lacks practical application validation, which needs further exploration to verify its applicability.
Experimental Evidence Dimension
In terms of experimental evidence, the authors constructed a DNA origami hinge model to validate the intercalator-driven rotational motion mechanism in detail. In the experiments, by varying the concentration of intercalators, a linear increase and saturation phenomenon in the hinge’s rotation angle were observed; by changing the types of intercalators (such as ethidium bromide EtBr and propidium iodide PR) and their mixing ratios, the sensitivity of hinge motion was regulated; and by adjusting the distance between AuNPs, the maximum rotation angle of the hinge was modulated. Additionally, by constructing curved nanoactuators, the potential for motion control of the hinge at the nanoscale was demonstrated. The experimental design is rigorous, employing various methods such as atomic force microscopy (AFM) imaging and SNUPI program simulations to validate the results, with experimental data supporting the precise control of the dynamic behavior of DNA origami hinges by intercalators.
However, the observed distribution of rotation angles was relatively wide, which may be due to non-uniform intercalator binding, affecting the accuracy and reproducibility of the experimental results. Future research could attempt to optimize the binding process of intercalators to achieve more uniform binding, thereby reducing the width of the rotation angle distribution.
Reasoning Analysis Dimension
The authors conducted reasonable logical reasoning based on the experimental results. For example, by observing the changes in the hinge’s rotation angle at different intercalator concentrations, they inferred that the binding of intercalators to DNA led to the twisting deformation of DNA bundles, thereby triggering the rotational motion of the hinge, and further proposed the idea of regulating the sensitivity and range of hinge motion by changing the types and concentrations of intercalators. The article also established a kinetic model to simulate the dynamic behavior of the hinge and compared the simulation results with experimental data, inferring the binding interactions of intercalators with the hinge in practical situations.
This model’s application not only aids in a deeper understanding of the system’s dynamic processes but also predicts the system’s behavior under different regulatory conditions, providing strong theoretical support for experimental design and result interpretation.
Video of Literature Sharing:
Original Link
https://pubs.acs.org/doi/10.1021/acsnano.4c09654
Written by: Cheng XianzhiProofread by: Ye XinchengPosted by: Ye Xincheng