Click the “Nanjing IFB” above to follow us!
Hello everyone, today I would like to share an article published inJACS titled:Uncovering the Molecular Landscape of Tetracycline Family Natural Products through Bacterial Genome Mining. The corresponding author is Researcher Guohui Pan from the Institute of Microbiology, Chinese Academy of Sciences.Researcher Guohui Pan’s research focuses on the discovery of microbial natural product drugs, elucidation of biosynthetic mechanisms, and synthetic biology research.
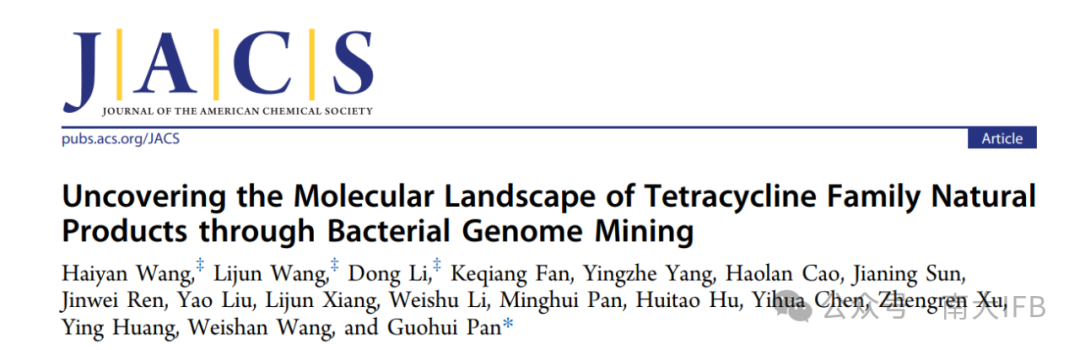
Tetracycline (TC) natural products are one of the most successful classes of antibiotics, having been in clinical use for over70 years, widely used to treat bacterial infections, parasitic diseases, and certain cancers. Tetracycline antibiotics exert their broad antibacterial activity by binding to the 30S subunit of the ribosome, blocking the binding of aminoacyl-tRNA to the A site, thereby inhibiting protein synthesis. However, most antibiotic molecules bind to similar or adjacent functional sites on the ribosome, and the emergence of rRNA modifications or drug-modifying enzymes leads to the inactivation of these sites, resulting in cross-resistance of bacteria to different types of antibiotics. To combat the spread of resistant bacteria and improve drug activity and stability, new tetracyclines are primarily obtained through semi-synthesis and total synthesis. However, in the past three decades, very few new tetracycline natural products have been discovered, thus exploring and developing novel tetracycline natural products will provide important resources for the development of a new generation of antibiotics and anticancer drugs.
The biosynthetic process of tetracycline compounds has been thoroughly studied, and the cyclization process of its skeleton is as follows:OxyK catalyzes the cyclization of C7-C12 to form the first ring,OxyN catalyzes the formation of rings 2 and 3, followed by the acyl-CoA ligaseOxyH converting ACP-thioester to AMP-thioester,OxyI catalyzes the hydroxyl-aldehyde condensation between C18 methylene and C1 carbonyl to complete the formation of the fourth ring. Among them, the fourth ring formation process catalyzed jointly byOxyH andOxyI is a key part that distinguishes it from the anthraquinone-type aromatic polyketone skeleton formation. Furthermore,OxyK,OxyN,OxyH, andOxyI homologs are conserved in the tetracycline biosynthetic gene cluster, andOxyH is significantly absent in other aromatic polyketones, thus the co-occurrence of these four proteins is a reliable marker for identifying tetracycline biosynthetic gene clusters.
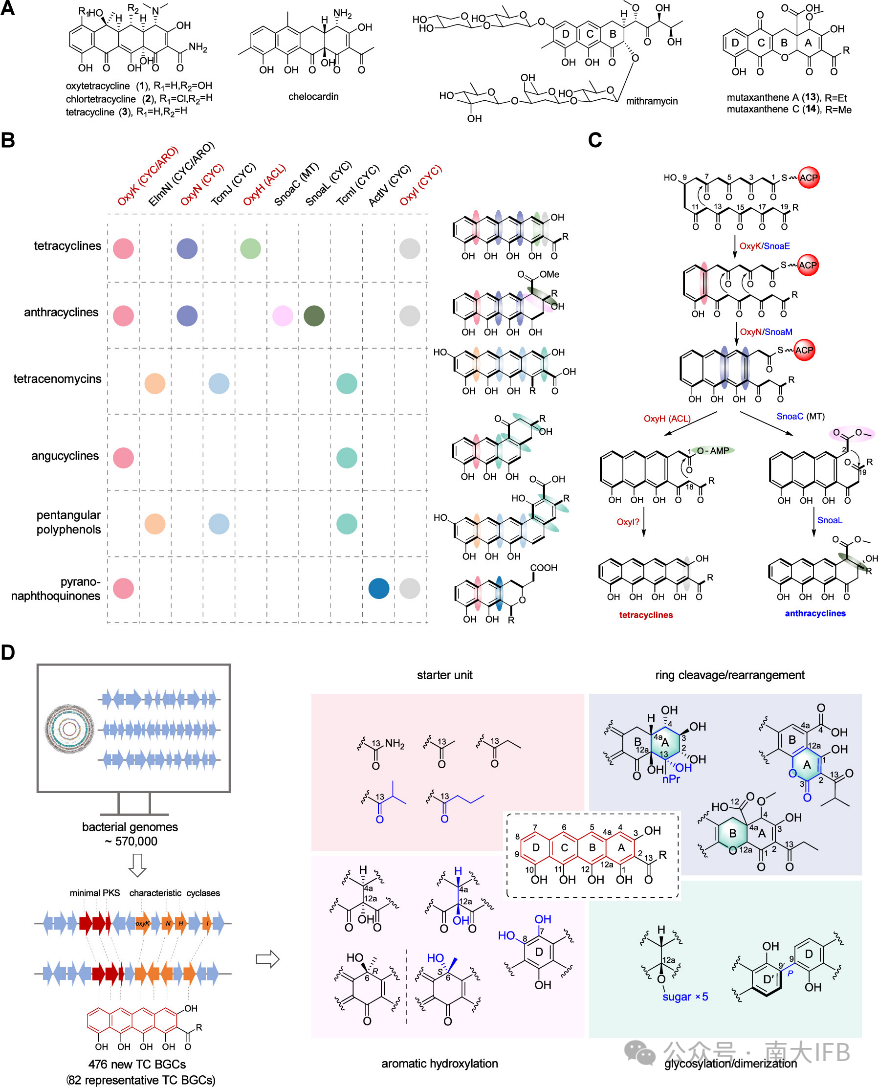
Figure 1. Discovery of novel tetracycline natural products through bacterial genome mining
First, the authors usedminimal PKS and tetracycline cyclases(OxyK, OxyN, OxyH, OxyI) as probes to mine476 new tetracyclineBGCs from the NCBI, JGI IMG/M bacterial genome databases. Further systematic comparison of the evolutionary and functional relationships of tetracyclineBGCs yielded82 representativeBGCs. These new representativeBGCs exhibited varying degrees of differences in gene composition and abundance, indicating their great potential in biosynthesis of structurally diverse novel tetracycline natural products.
To further support the bioinformatics analysis and identify other potential producing strains, the authors selected several clusters from known compounds (such as soil mycin, tetracycline,Mutaxanthenes) branches for exploration, all of which produced corresponding known compounds, validating the accuracy of the analysis and enriching the natural producing strains of soil mycin, tetracycline,Mutaxanthenes, which may provide valuable genetic elements and strain resources for developing high-yield strains of these compounds.
Next, the authors studied three gene clusters rich in post-modification genes. First, overexpression of the transcriptional regulatorSARP activated theStreptomyces misionensis DSM 40306 mis gene cluster, leading to the discovery of novel tetracycline compounds with different glycosylation modificationsmisiomycin A-D. They are characterized by a common tetracycline core structure, with a special isobutyryl group at the C2 position as the starting unit. Among them,misiomycin A has the most complex glycosylation modifications, with the longest and most complex sugar chain at the C12a position. The order of action of glycosyltransferases was determined by knocking out glycosyltransferases (Figure 2).
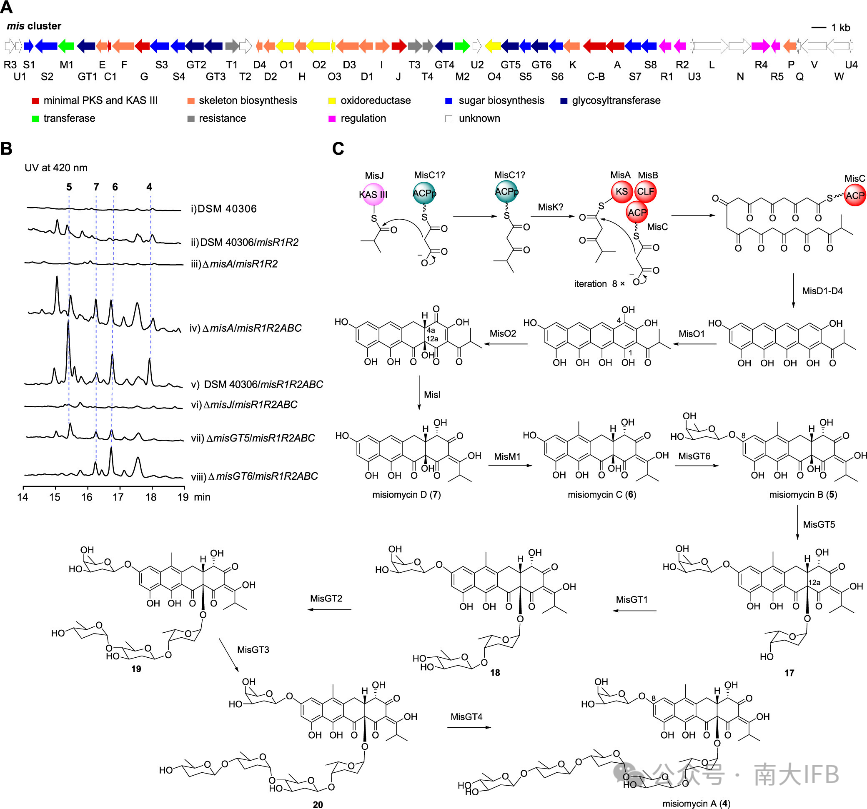
Figure 2. Discovery of Misiomycin and proposed biosynthetic pathway
Streptomyces varsoviensis NRRL ISP-5346 contains two tetracycline gene clusters, and overexpression of the regulatory factor on thevar cluster led to the production of varsomycin A andB, whose structures were determined by NMR analysis and X-ray single crystal diffraction. They are characterized by a special isobutyryl group at the C2 position, wherevarsomycin A consists of a pair of enantiomers, and also features a novel six-membered lactone ring structure formed by C3-O-C12. Through structural analysis and corresponding gene knockout validation, it was found thatvarsomycin is a product of the synergistic action of two tetracycline biosynthetic gene clusters in this strain,var andoxt, utilizing isobutyryl-CoA as the starting unit and recruiting enzymes from theoxt cluster, includingC9-ketoreductaseOxtJ andC6-methyltransferaseOxtF (Figure 3).
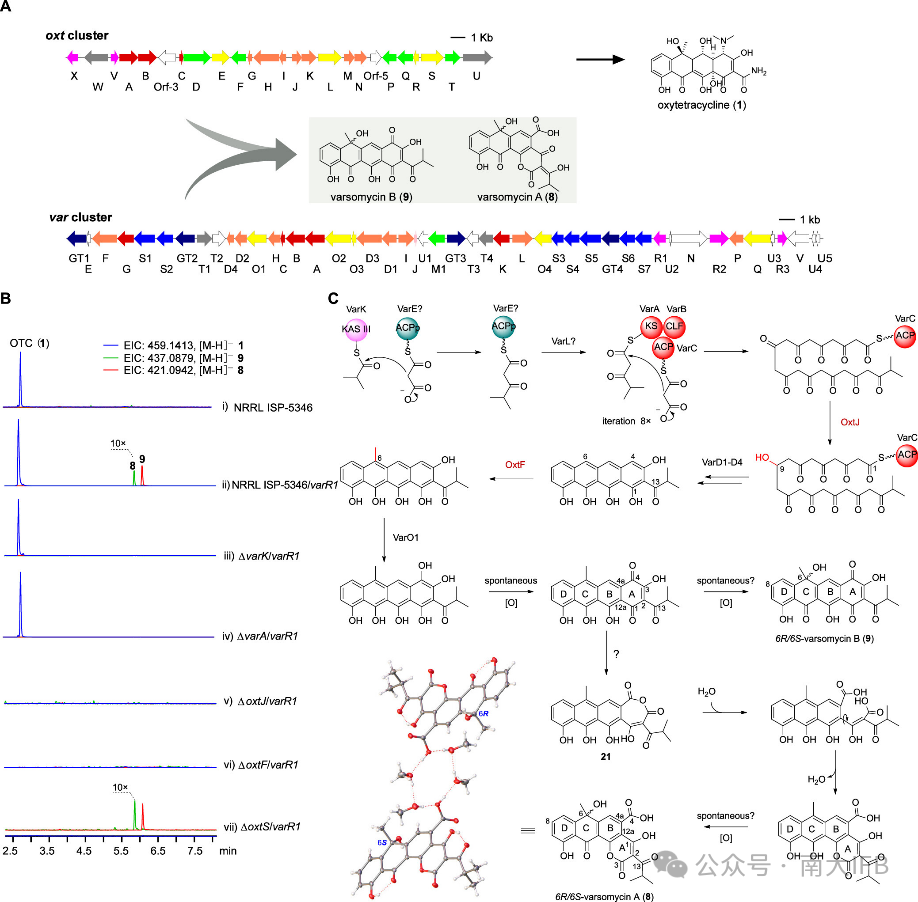
Figure 3. Discovery of Varsomycin and proposed biosynthetic pathway
Saccharothrix australiensis DSM 43800 contains thehib gene cluster rich in FAD-dependent oxidoreductases. Compoundshibarimicins J–L were isolated and identified from the fermentation broth of this strain, and feeding experiments using monomeric compound24 indicated that compound24 is a biosynthetic precursor of dimeric products. Subsequently, based on gene knockout and the enzymatic reaction results of radical SAM enzymes, the biosynthetic process ofhibarimicins J–L was proposed (Figure 4).
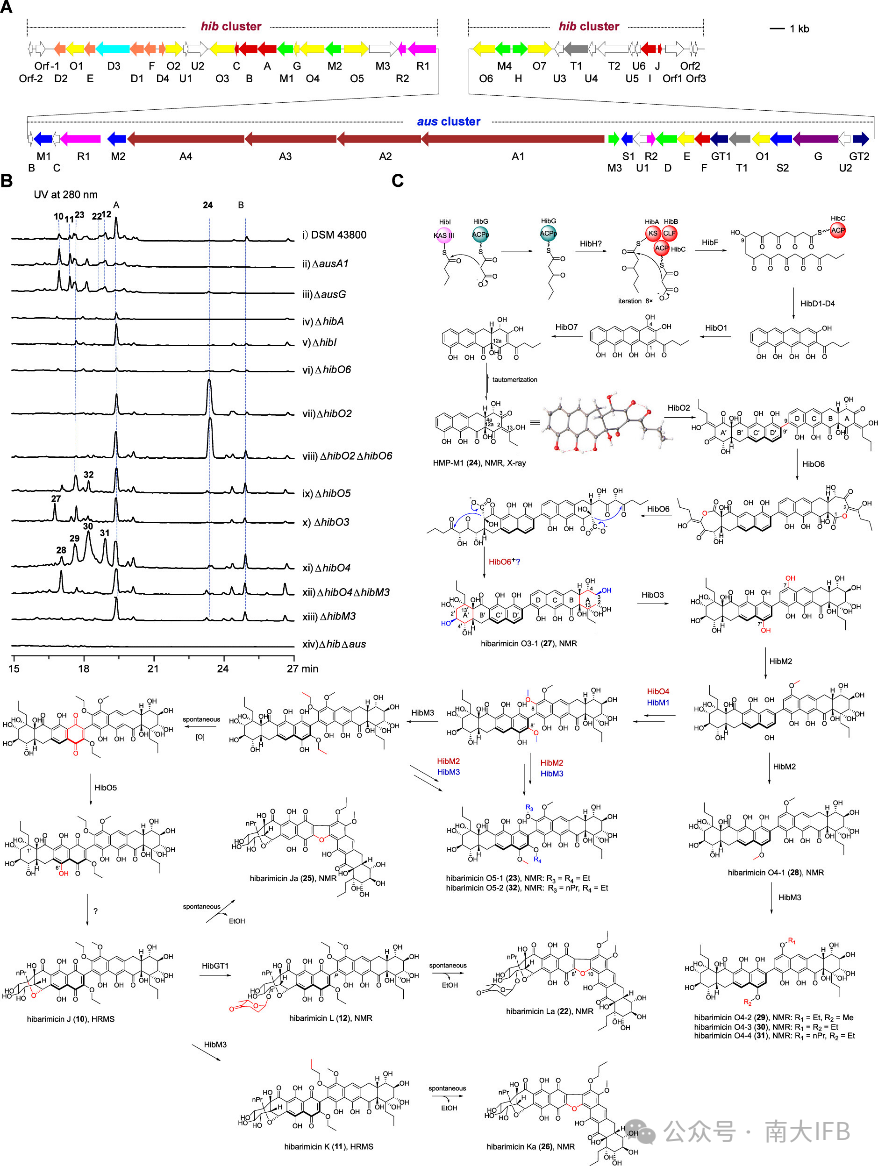
Figure 4. Discovery of Hibarimicins and proposed biosynthetic pathway
Finally, the authors conducted antibacterial and anticancer activity assays on the isolated compounds (Table 1). Misiomycin A exhibited strong antibacterial activity against vancomycin-resistant Enterococcus faecium (VREfm) V583, methicillin-resistant Staphylococcus aureus (MRSA ) 113, and Bacillus subtilis 168, with MIC values of 1-3 μM. Compound 24 showed significant antibacterial activity against Staphylococcus aureus ATCC 6538, MRSA and VREfm, with MIC ranges of 2-4 μM. Hibarimicins L exhibited effective antibacterial activity against Staphylococcus aureus ATCC 6538 and MRSA, with MIC values of 0.5 and 1 μM..
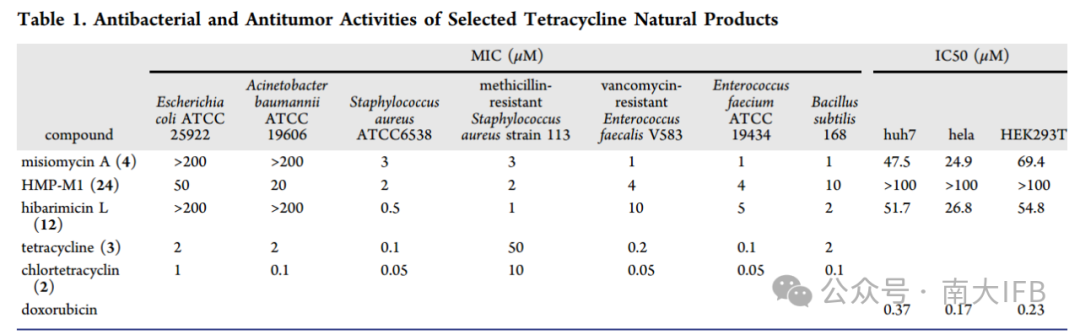
Table 1. Antibacterial and anticancer activities of tetracycline natural products
Summary: This study developed a precise strategy based on specific cyclases to discover tetracycline biosynthetic gene clusters, thereby identifying82 representative gene clusters with the potential to produce structurally diverse tetracycline natural products, from which three groups of novel natural products were identified, revealing their biosynthetic processes. These compounds exhibit various unique structural features, including special starting units, complex oxidative rearrangements, monomer dimerization, and complex glycosylation patterns, expanding the chemical diversity of tetracycline family natural products and providing important resources for the development of a new generation of antibiotics and anticancer drugs.
Article link: https://doi.org/10.1021/jacs.4c17551
Article title: Uncovering the Molecular Landscape of Tetracycline Family Natural Products through Bacterial Genome Mining
Organizer: Wenfei Tan
Long press the image below to recognize the QR code in the image and easily follow us!
