Cancer, as a highly heterogeneous disease, involves complex changes across multiple layers, including the genome, epigenome, transcriptome, proteome, and metabolome. In recent years, with the rapid development of high-throughput sequencing technologies, the integrated analysis of multi-omics data has provided unprecedented opportunities for cancer research. Deep learning (DL) technology can analyze high-dimensional datasets to discover new disease mechanisms and biomarkers, aiding in the improvement of patient treatment and management. A recent review article published in the journal Genes systematically reviews the latest advancements in deep learning and multi-omics in cancer research, focusing on four core application areas: cancer type/subtype classification, driver gene prediction, survival analysis, and drug response prediction, demonstrating the significant role of the integration of deep learning and multi-omics data in enhancing our understanding of biological systems, particularly in cancer research.
Significance of Integrating Deep Learning and Multi-Omics
Traditional cancer research often limits itself to a single omics layer, while modern studies indicate that the occurrence and development of cancer result from the interplay of various molecular mechanisms. Deep learning, as an important branch of machine learning, has become a key technology for integrating multi-omics data and revealing the complex mechanisms of cancer due to its powerful nonlinear modeling capabilities and automatic feature extraction advantages.
The literature points out that deep learning has shown unique value in analyzing high-dimensional genomic data, capable of discovering disease mechanisms and biomarkers that traditional methods struggle to capture. Particularly in cancer research, deep learning architectures such as Convolutional Neural Networks (CNN), Graph Neural Networks (GNN), Recurrent Neural Networks (RNN), and Transformer-based networks have been successfully applied in several key areas, including differential gene expression classification, patient stratification based on molecular features, survival analysis prediction, and somatic mutation identification.
Notably, deep learning has also made significant progress in the analysis of histopathological images. Recent studies show that tumor tissue morphological features extracted through deep learning can be associated with molecular features (such as tumor mutation burden, RNA expression, etc.), providing a cost-effective alternative for clinical practice, potentially replacing whole-genome sequencing (WGS) and whole-exome sequencing (WES) technologies.
Applications of Deep Learning and Multi-Omics Integration
1. Classification of Tumor Types, Subtypes, and Cancer of Unknown Primary
Molecular typing is the cornerstone of precision oncology, allowing for cancer classification based on molecular features rather than solely on histopathology. The literature details various innovative deep learning models for molecular typing classification tasks, such as:
- DeepType: By integrating supervised and unsupervised learning, it transforms gene expression data into latent space representations with better separability, successfully identifying molecular subtypes significantly associated with clinical prognosis in breast cancer and bladder cancer data.
- Subtype-GAN: A multi-input, multi-output framework based on a Generative Adversarial Network (GAN) that can simultaneously handle various omics data types (copy number, DNA methylation, miRNA, and mRNA data, etc.). Its ability to identify five known breast cancer subtypes was validated across ten tumor types based on the TCGA database.
- MuAt: A deep neural network based on attention mechanisms that predicts 24 tumor types using whole-genome and whole-exome sequencing data, achieving accuracies of 89% and 64%, respectively. Its innovation lies in its ability to identify unannotated tumor subtypes and associate them with known mutation features. These tools provide significant support for clinical decision-making, especially in healthcare environments with limited molecular testing resources.
2. Prediction of Cancer Driver Genes
Identifying driver genes is crucial for understanding the mechanisms of cancer onset, assessing patient prognosis, and developing targeted therapies. The literature compares the limitations of traditional methods (such as frequency-based and network-based approaches) and highlights various innovative deep learning solutions, such as:
- FI-Net: Predicts driver genes through Functional Impact Scores (FIS), validated across 31 TCGA cancer types, with 53% of predicted genes consistent with known cancer gene databases (CGC).
- EMOGI: An interpretable graph convolutional network model that integrates multi-omics data and protein interaction networks, achieving an average AUPRC of 71%. These models can not only identify known driver genes but also discover new candidate genes, such as the 165 potential new cancer genes (NPCGs) identified by EMOGI, providing new directions for cancer biology research and targeted therapy development.
3. Multi-Omics Survival Analysis
Survival analysis is an important tool for studying biomedical time-event outcomes. In the context of multi-omics, deep learning models can integrate molecular features with clinical outcomes to achieve more accurate prognostic predictions:
- DeepOmix: Integrates gene expression, DNA methylation, copy number variation, and gene mutation data, achieving an average C-index of 0.69 across eight TCGA tumor types.
- MMOSurv: A meta-learning framework that can achieve effective predictions (C-index 0.67) in rare cancers with only 10-20 samples.
- PORPOISE: The first panoramic cancer prognosis model integrating histopathological images and genomic data, with an average C-index of 0.64. These models not only improve prediction accuracy but also enhance the interpretability of results through visualization techniques (such as attention heatmaps), providing reliable evidence for clinical decision-making.
4. Prediction of Drug Responses
Drug response prediction is a core challenge in precision oncology. The literature reviews major pharmacogenomic databases (such as CCLE, GDSC, CTRP, and PRISM) and various deep learning models. It particularly emphasizes the importance of evaluation strategies, noting the need to distinguish between different testing scenarios with varying degrees of rigor, such as random splits, unseen cell lines, unseen drugs, and unseen cell line-drug combinations, to ensure model generalization in the real world.
Challenges and Future Directions
Despite the immense potential of deep learning in multi-omics cancer research, the literature also points out current challenges:
- Standardization and Benchmarking: As multi-omics and deep learning become more prevalent, establishing unified standard practices and benchmarking standards becomes crucial, helping researchers select appropriate tools based on specific needs.
- Interpretability: Model transparency is a key prerequisite for clinical translation. Clinical applicability should always be considered when developing new methods; only when the workings of these models are fully understood and their reliability and robustness ensured can they be gradually integrated into clinical workflows.
- Data Bias: Biases present in biological data (such as limited genetic diversity in reference databases and insufficient representation of rare diseases) may lead to inaccurate model predictions in minority populations. The article suggests employing fairness algorithms (such as adversarial debiasing) and cross-group validation studies to mitigate this issue.
- Computational Costs: Particularly large language models based on Transformers (such as GenomeBert) face challenges of high computational resource demands, which may limit their application in resource-constrained environments.
Future development directions include:
- Developing more efficient and transparent model architectures
- Strengthening multi-center collaborations to build more representative datasets
- Promoting clinical translational research to validate model utility in real healthcare environments
- Exploring privacy-preserving technologies such as federated learning in multi-center studies
Conclusion
This review comprehensively outlines the current applications of deep learning in cancer genomics, particularly focusing on four major areas: cancer type classification, driver gene identification, drug response prediction, and survival analysis. With continuous technological advancements and the accumulation of clinical validations, deep learning-driven multi-omics analysis is expected to bring revolutionary changes to cancer precision medicine, ultimately achieving personalized treatment strategies tailored to individual patients. However, the translation of deep learning technology from the laboratory to the clinic will still require time; only when these models are fully understood and proven reliable can they be gradually integrated into routine clinical practice. Future research should aim to address current challenges and promote the field towards more standardized, transparent, and equitable directions.
References:
1. Sartori, F.; Codicè, F.; Caranzano, I.; Rollo, C.; Birolo, G.; Fariselli, P.; Pancotti, C. A Comprehensive Review of Deep Learning Applications with Multi-Omics Data in Cancer Research. Genes 2025, 16, 648. https://doi.org/10.3390/genes16060648
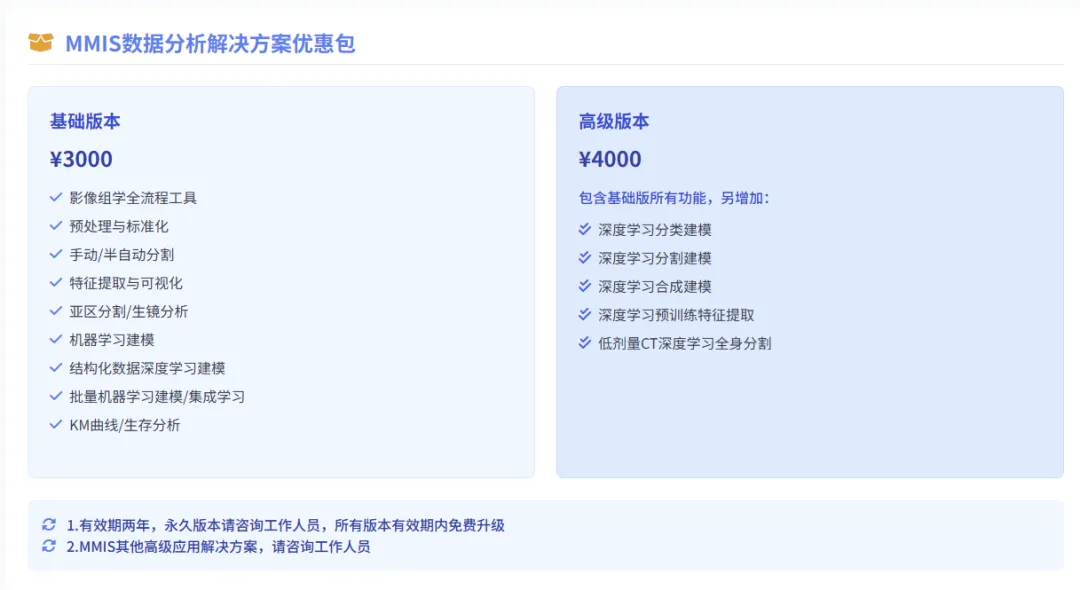
MMIS Service Content


Recommended Reading• Introduction to the MMIS Molecular Imaging Processing Platform
- Review and Outlook on 30 Years of Development of Nuclear Medicine Molecular Imaging Technology
- Introduction to MMIS Data Processing and Consulting Services
- Overview of Multimodal Fusion Methods for Image and Non-Image Data
- Introduction to Multimodal Data Fusion Methods
- Multimodal Imaging + Habitat Analysis: A Step-by-Step Guide to Habitat Partitioning
- 99mTc-MDP SPECT/CT Whole-Body Bone Scanning – Expected to Become a Routine Clinical Scanning Item
- Will SPECT Whole-Body Bone Scanning Replace Plain Bone Scanning?
- How to Publish an SCI Article: Practical Processes and Key Ideas
Previous Series Articles• Live Imaging Analysis• Metabolic Network Analysis• Advanced Applications of MMIS• MMIS Data Analysis Functions• Radiomics and Deep LearningDisclaimer: This shared content is for reference only and does not constitute any medical advice. The information and results provided in the article are not for actual clinical application but for preclinical research only! We do not assume responsibility for any direct or indirect losses! Reprint with acknowledgment: Source fromMMISMolecular Imaging!