


Introduction:
As of now, Nature has published a total of 572 articles. This article selects some of the latest literature to summarize color matching, providing five descriptive models for each color: hexadecimal, RGB, HSB, CMYK, and LAB, to facilitate future color matching usage.
Three Colors:
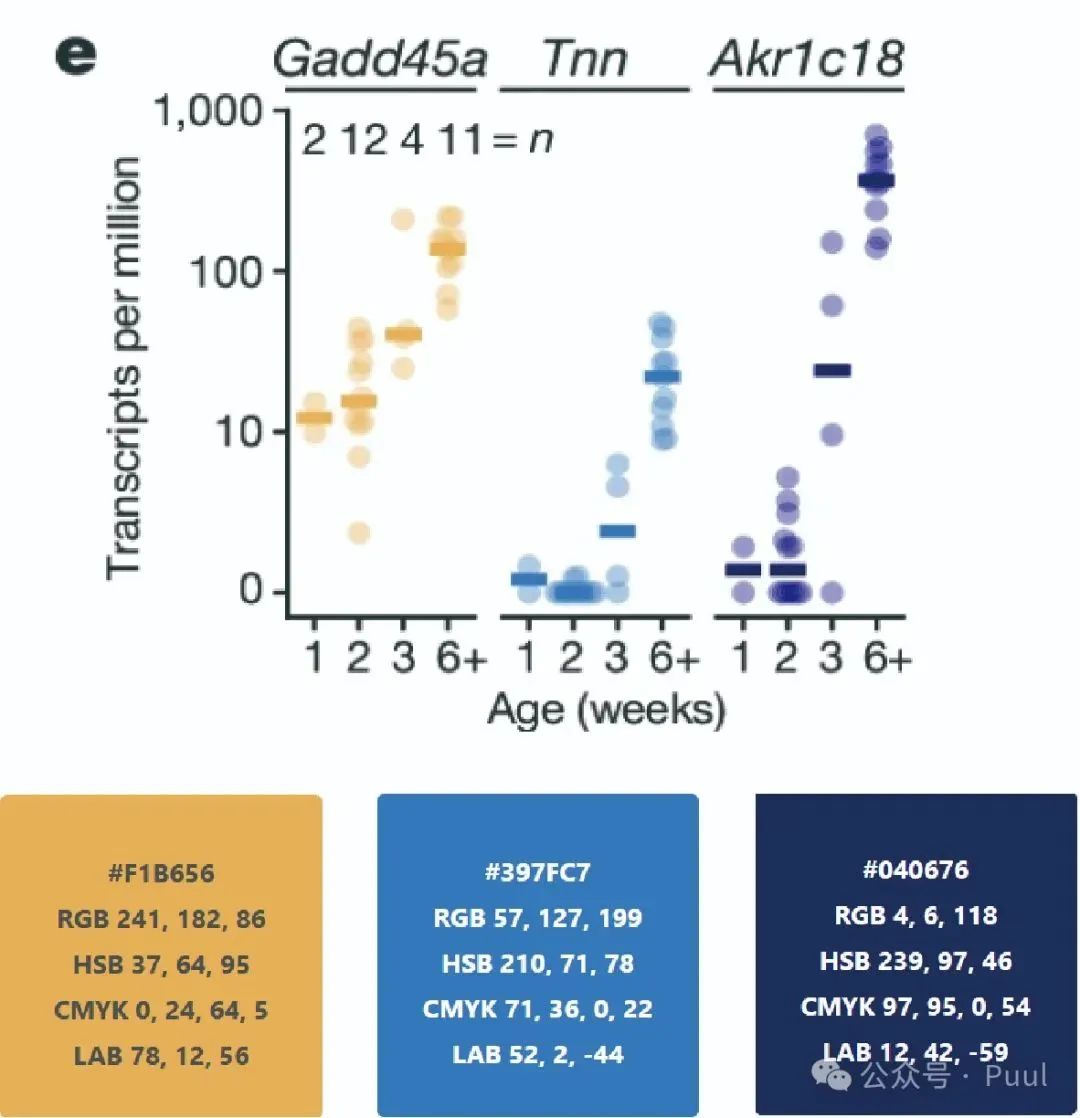
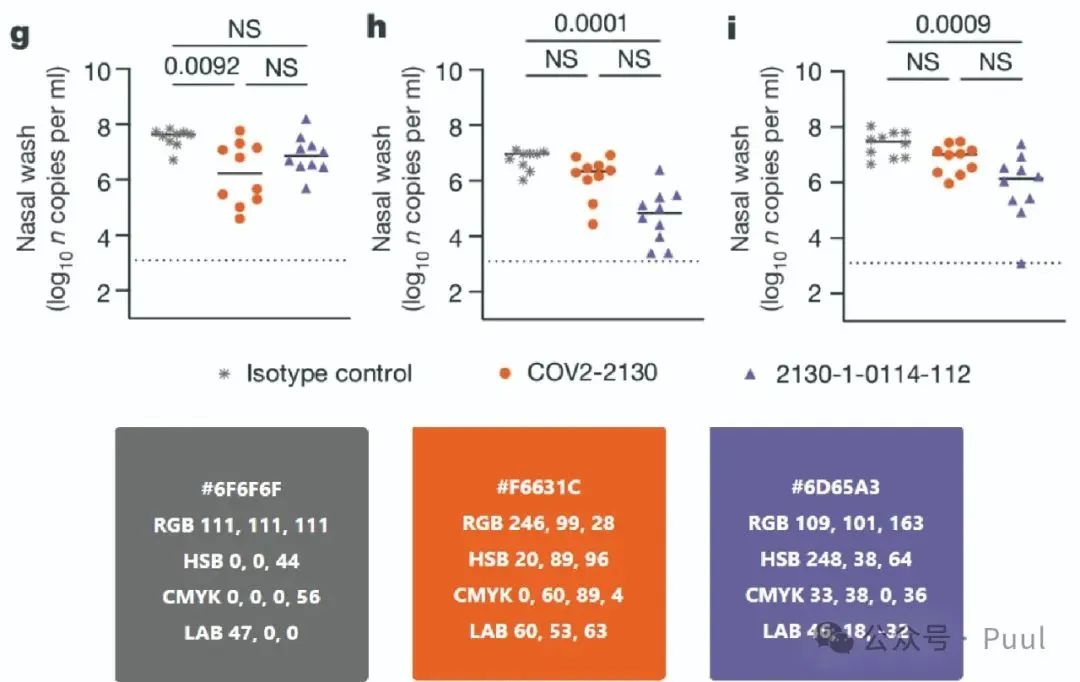
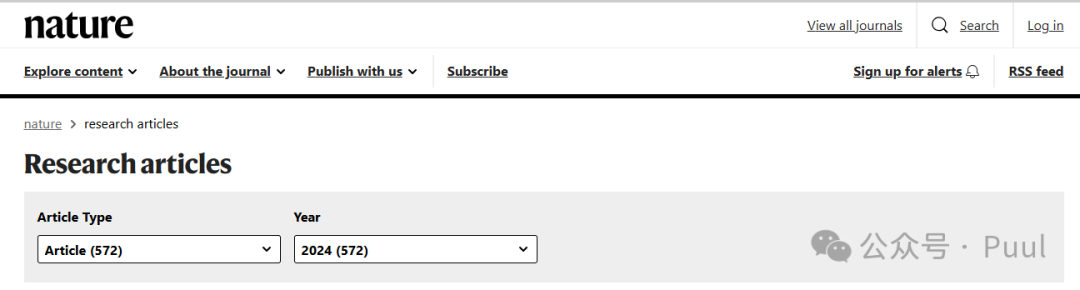
Four Colors:
Five Colors:
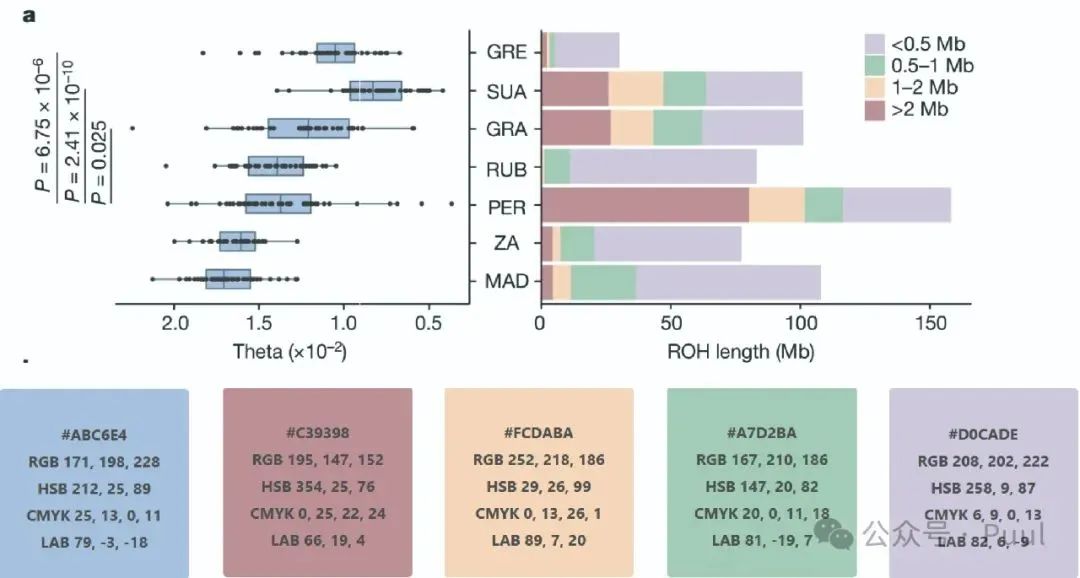
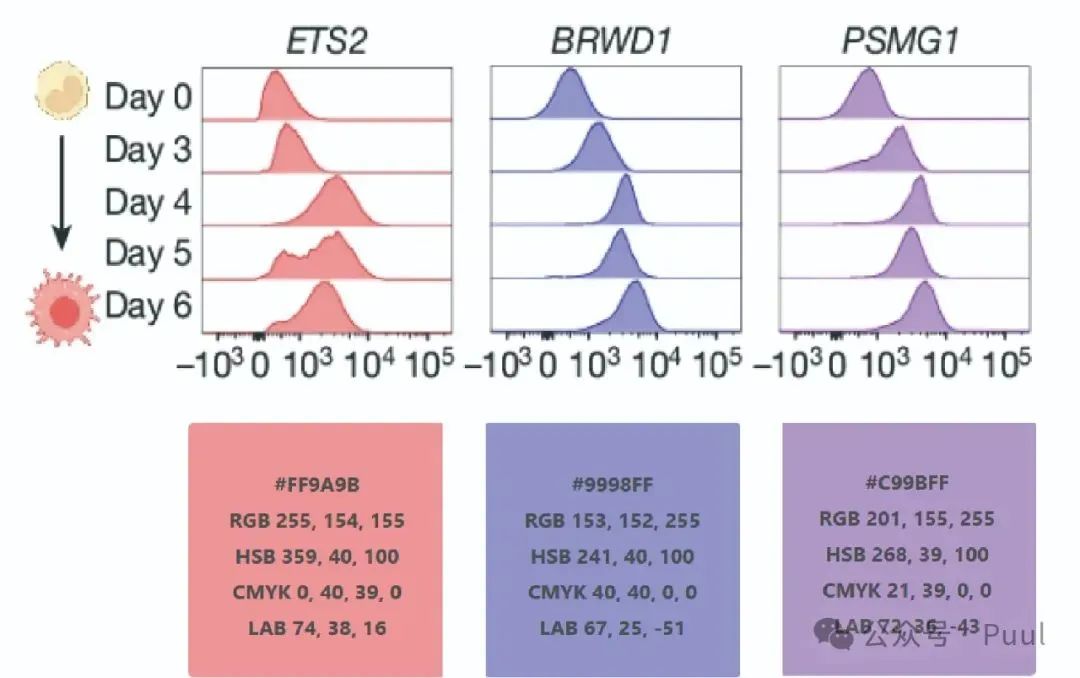
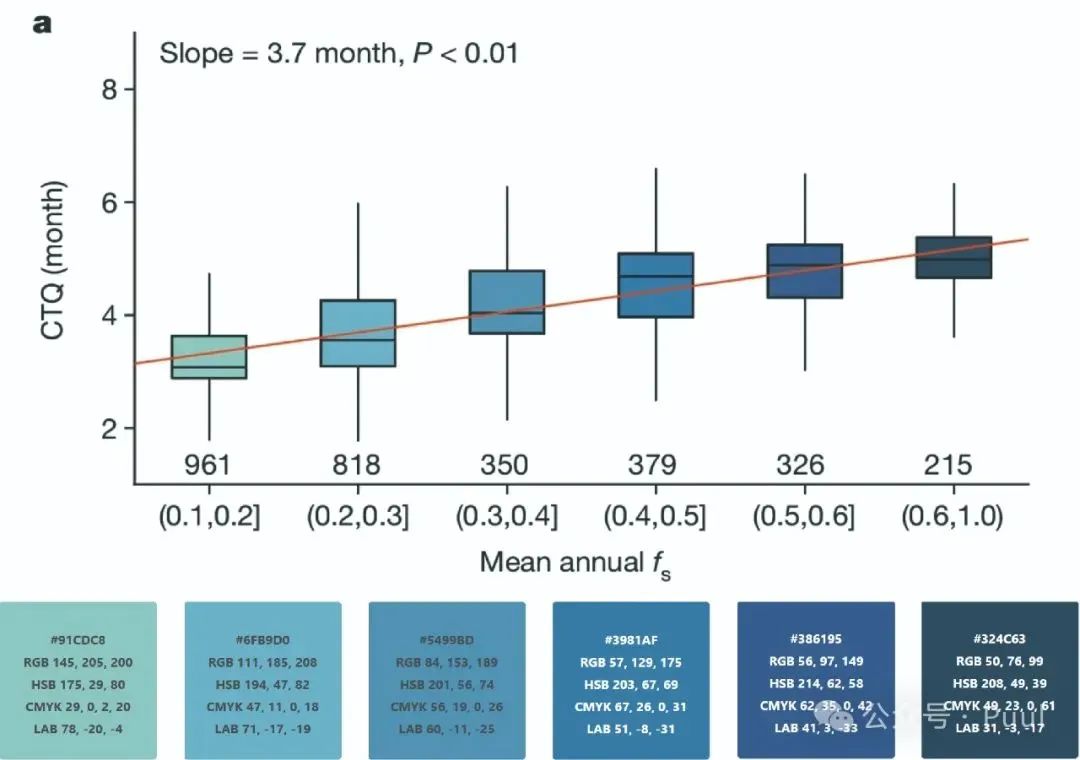
Six Colors:
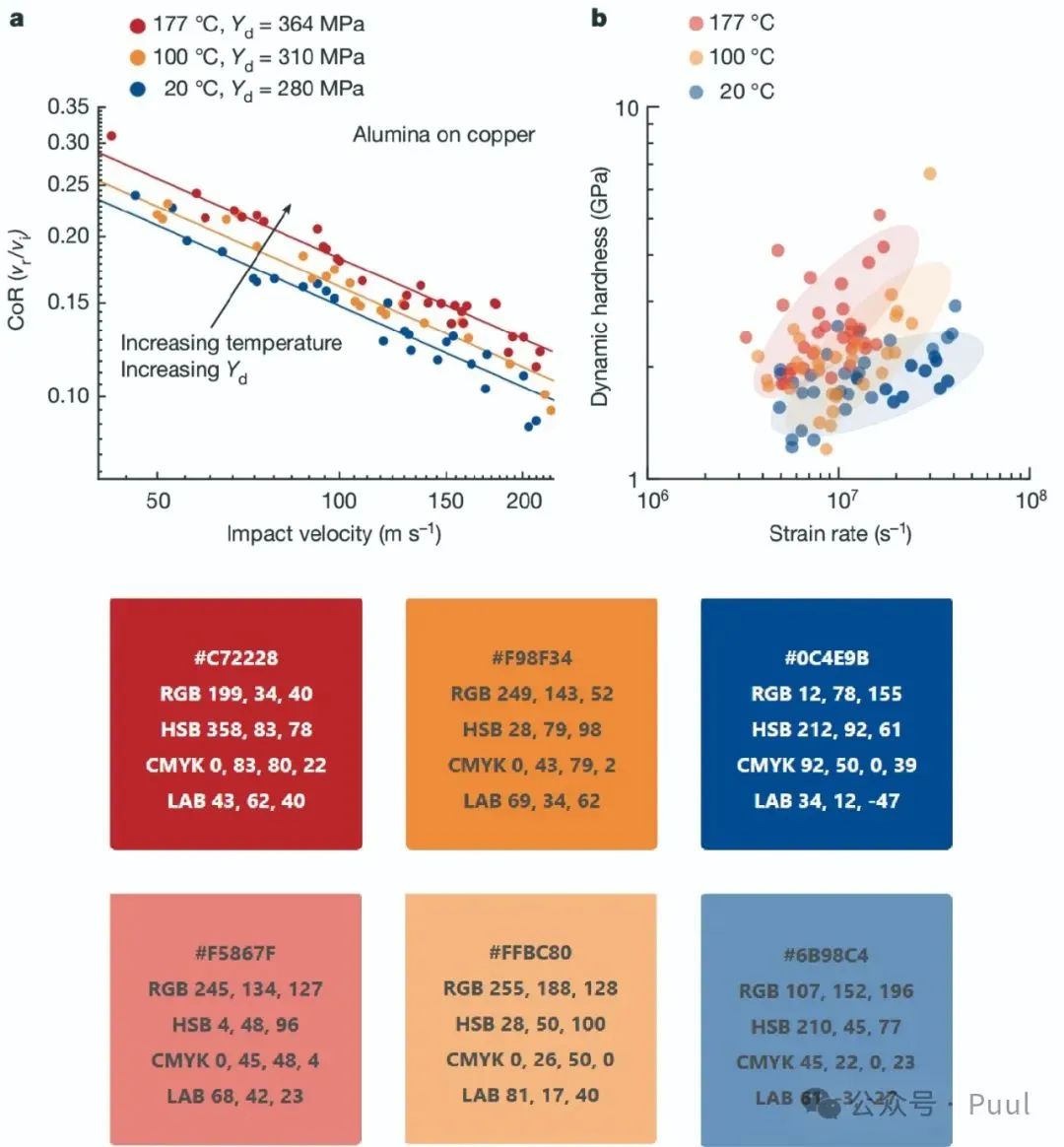
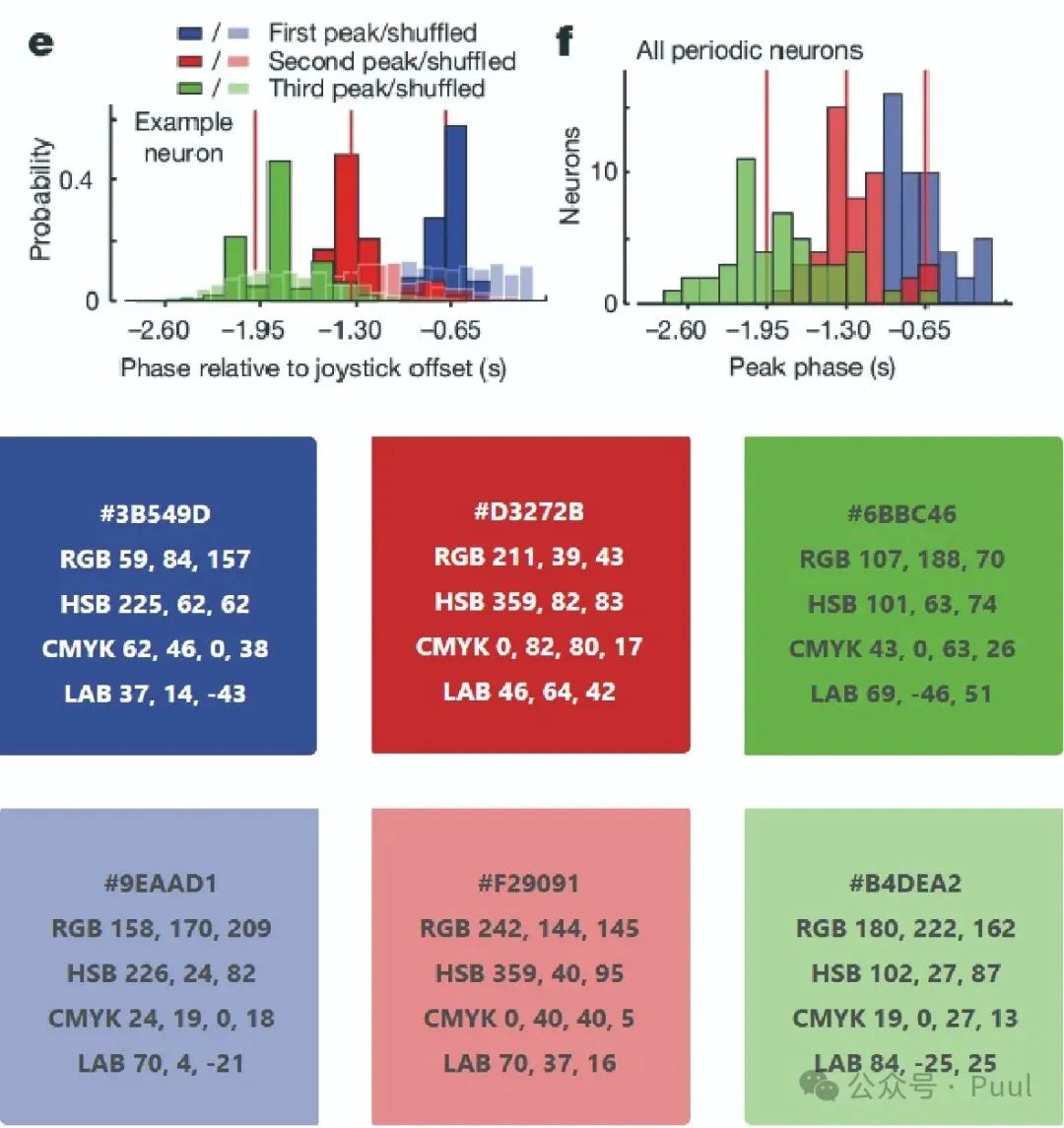
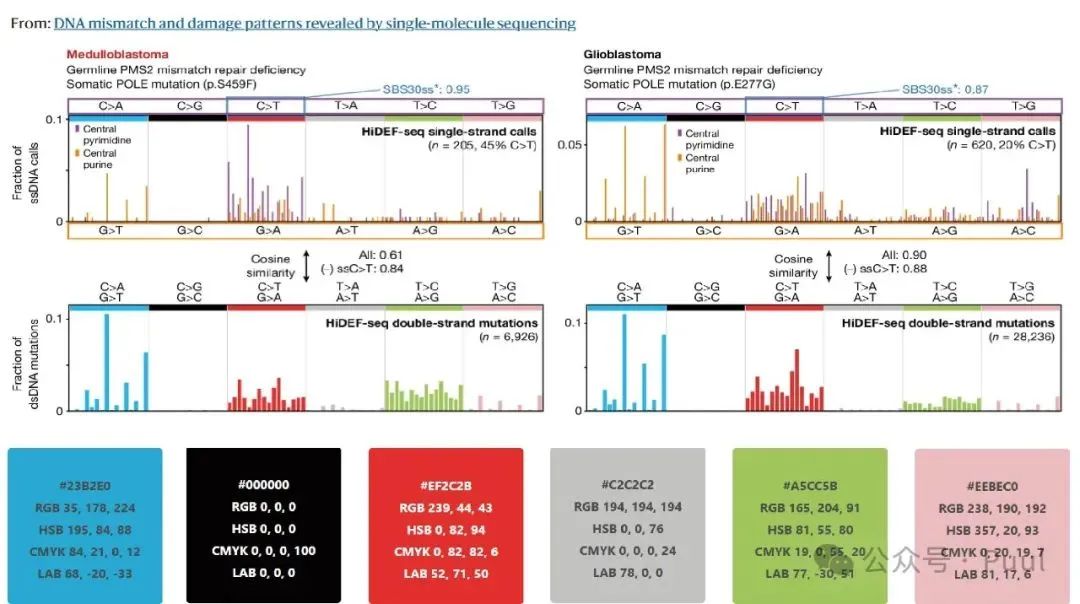
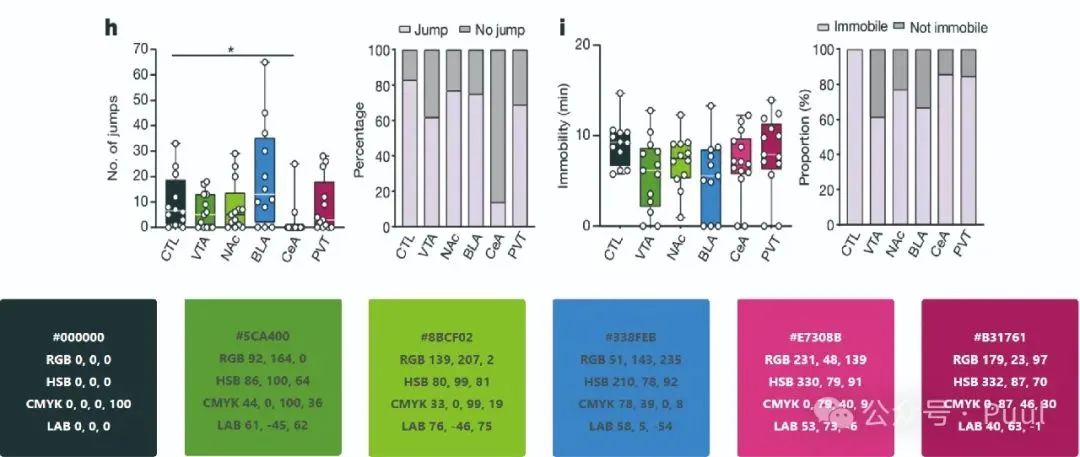
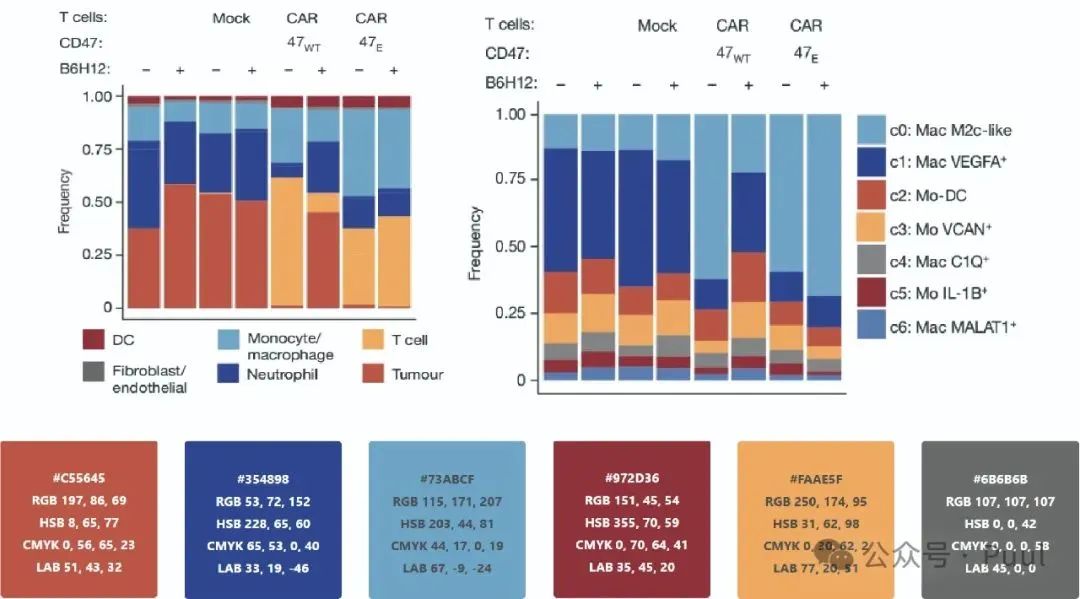
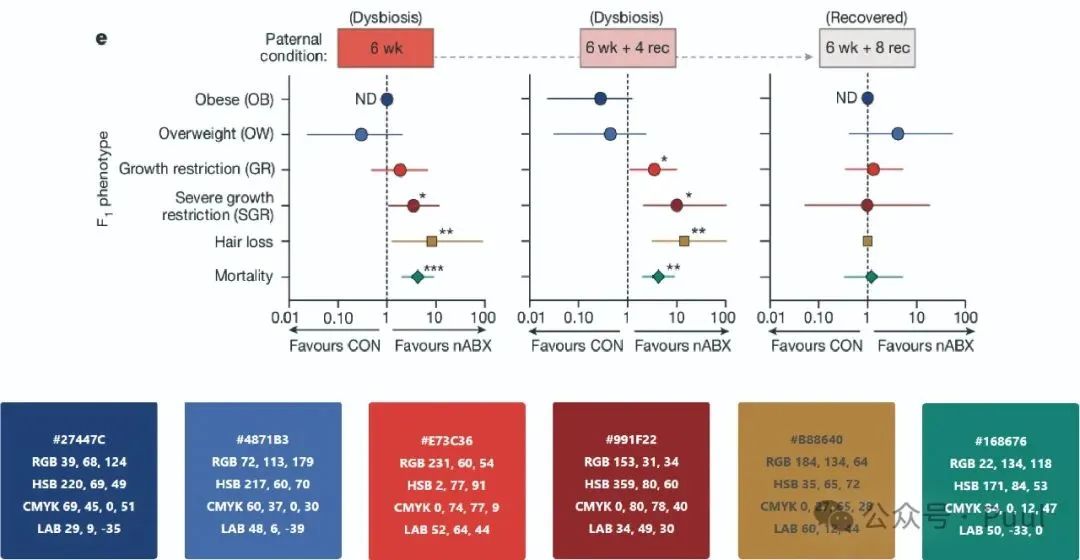
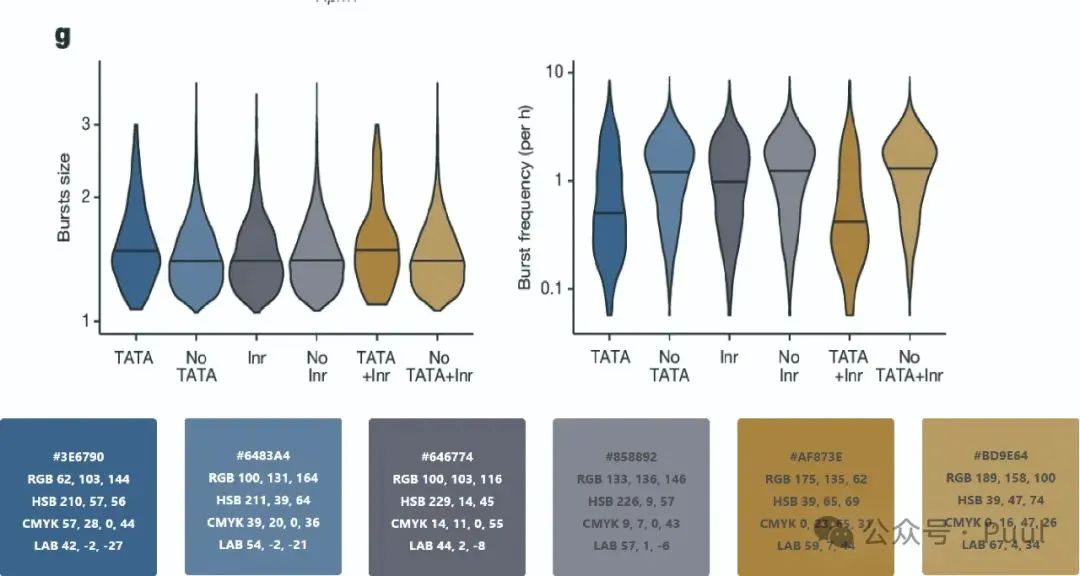
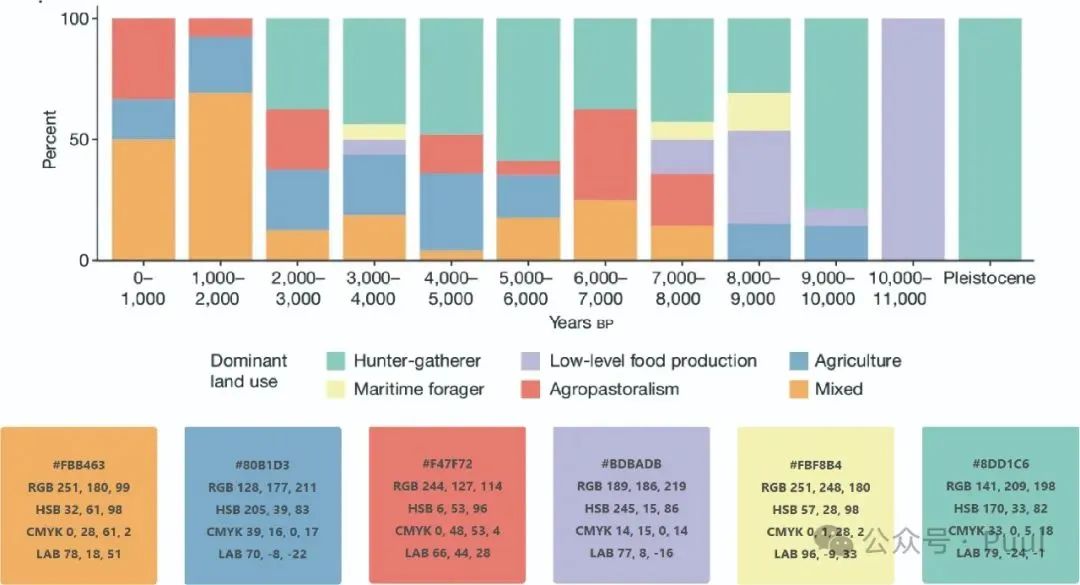
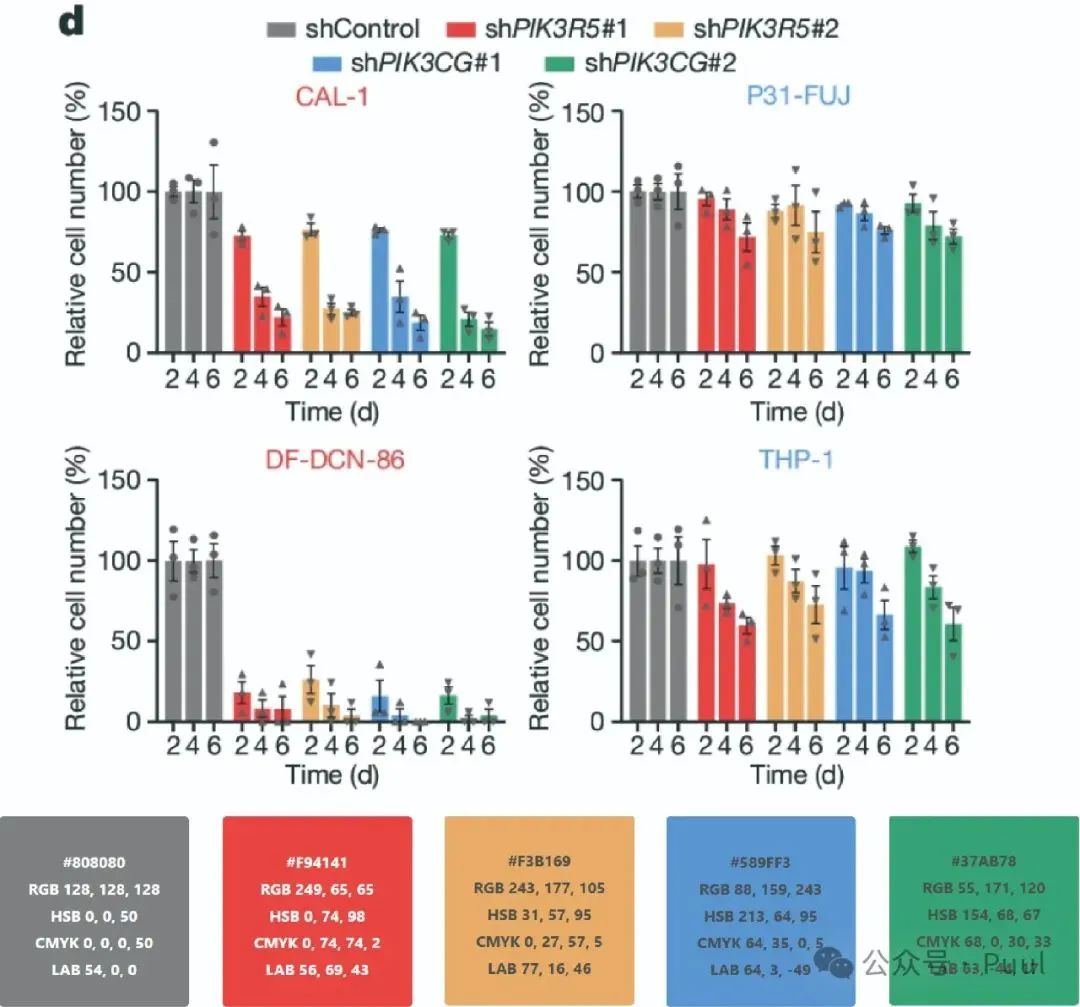
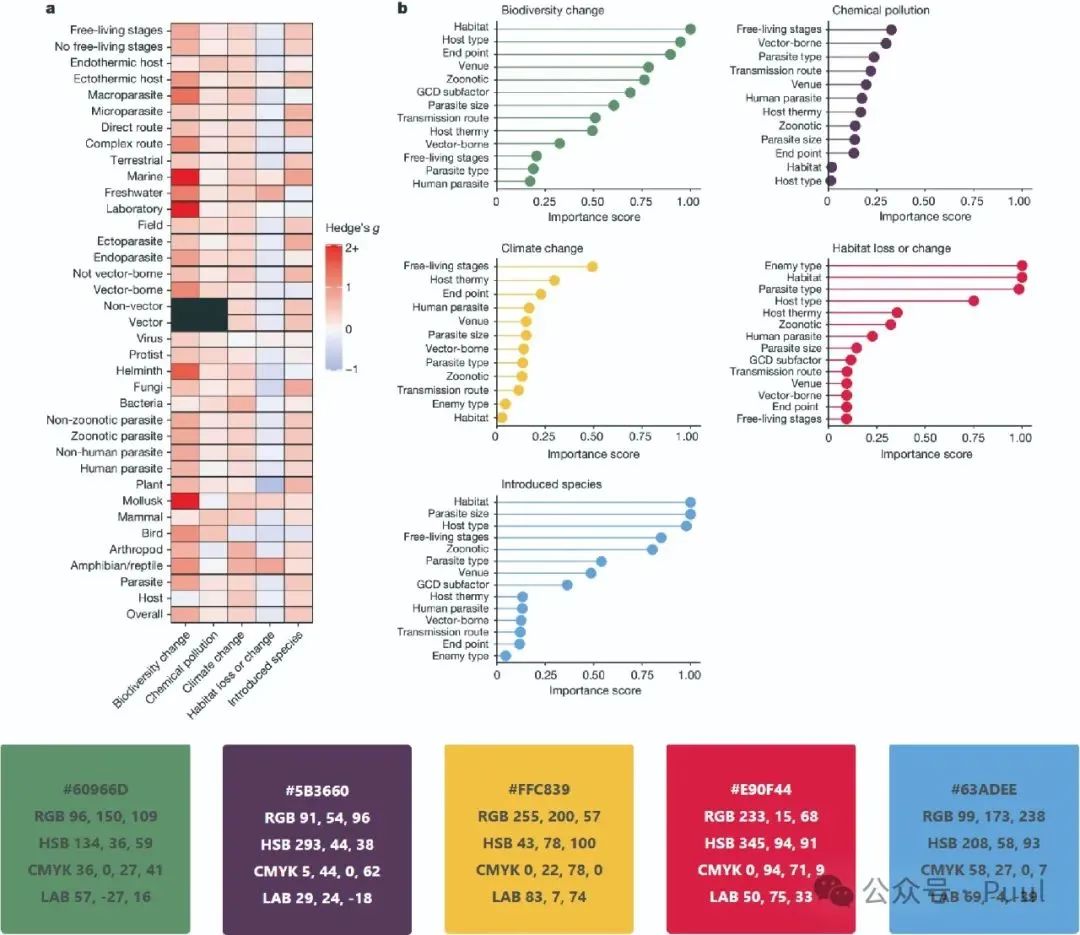
Multiple Colors:
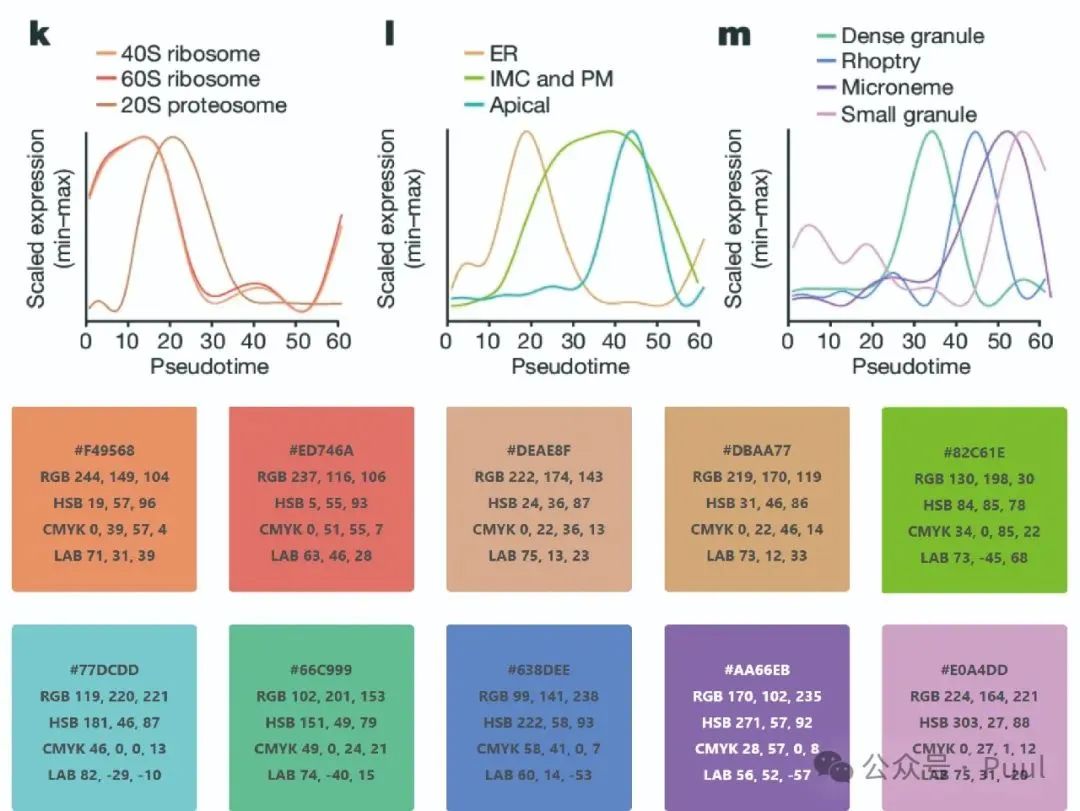
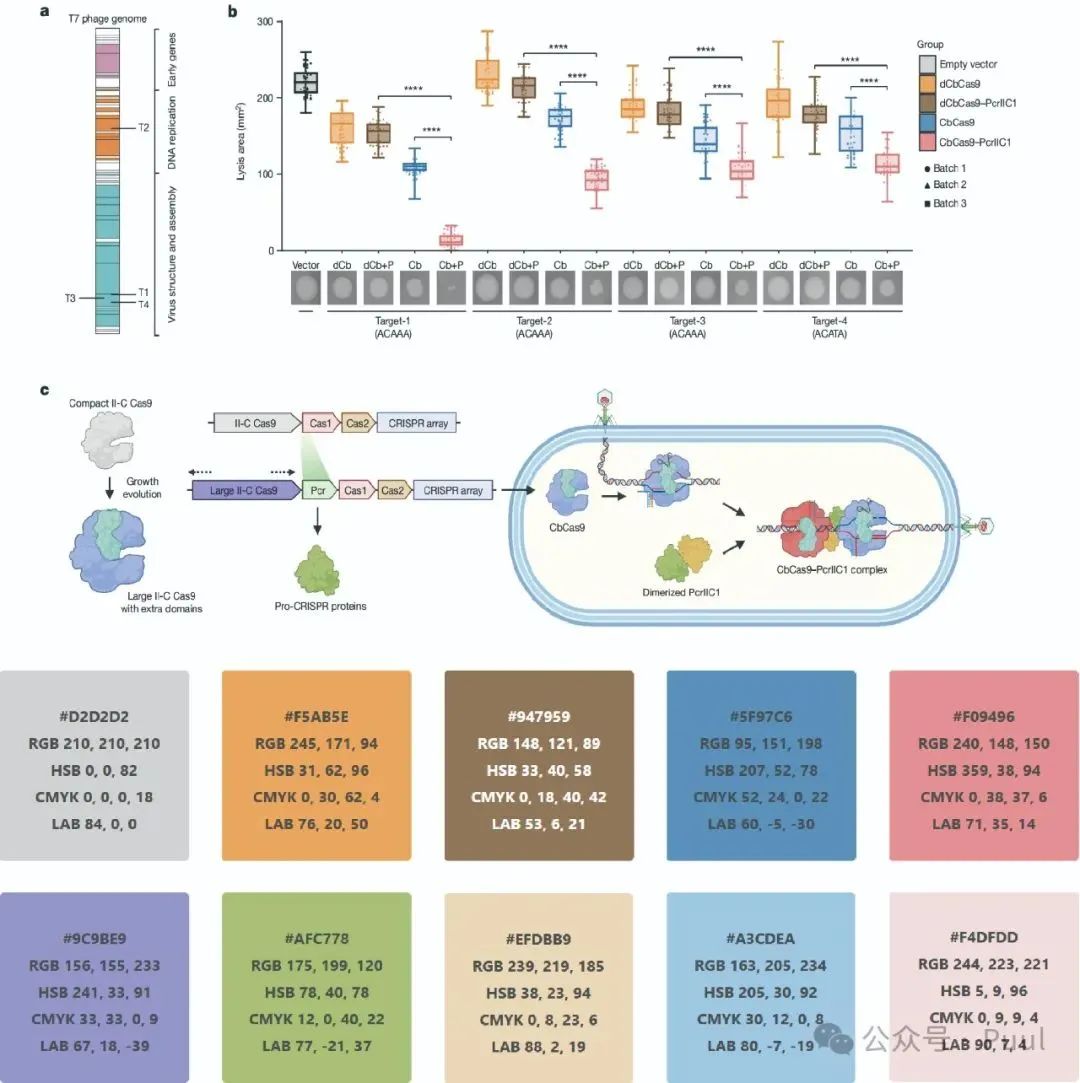
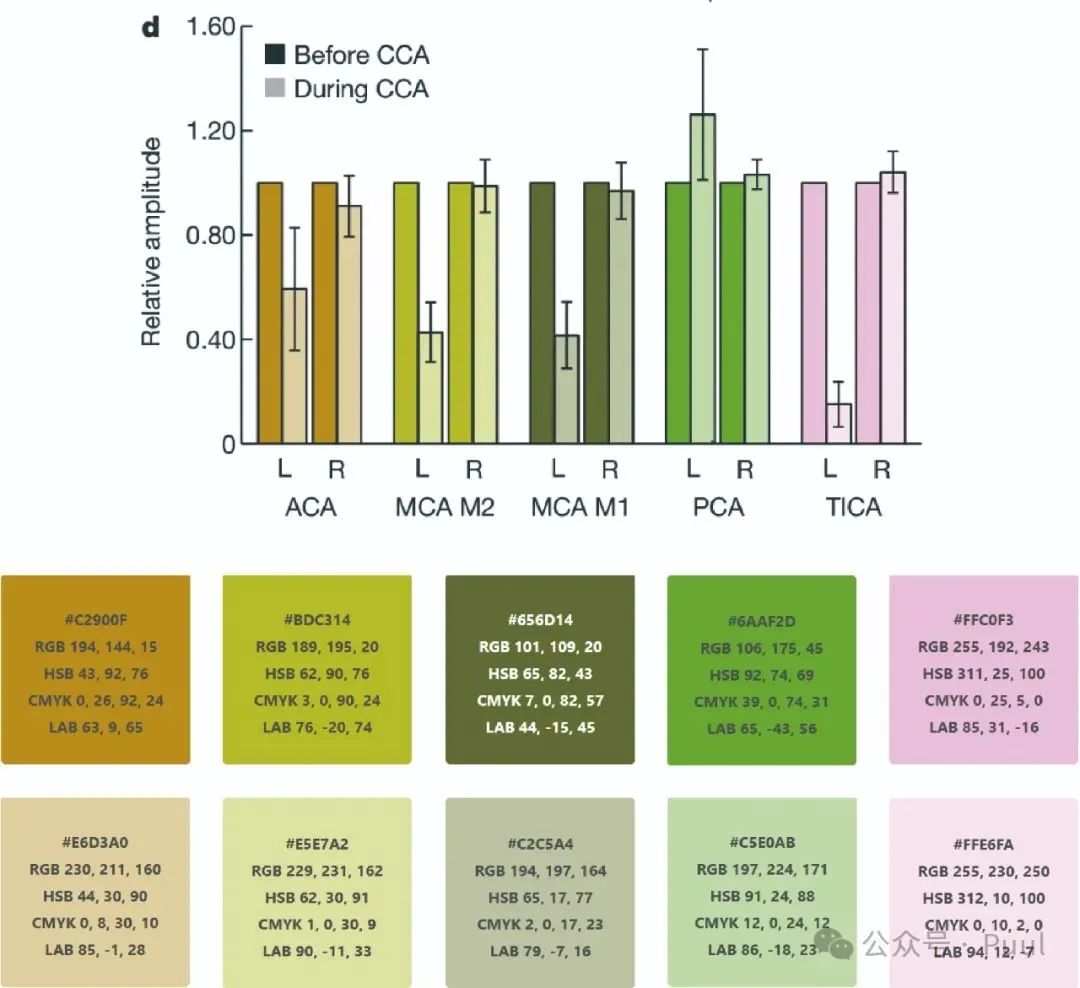
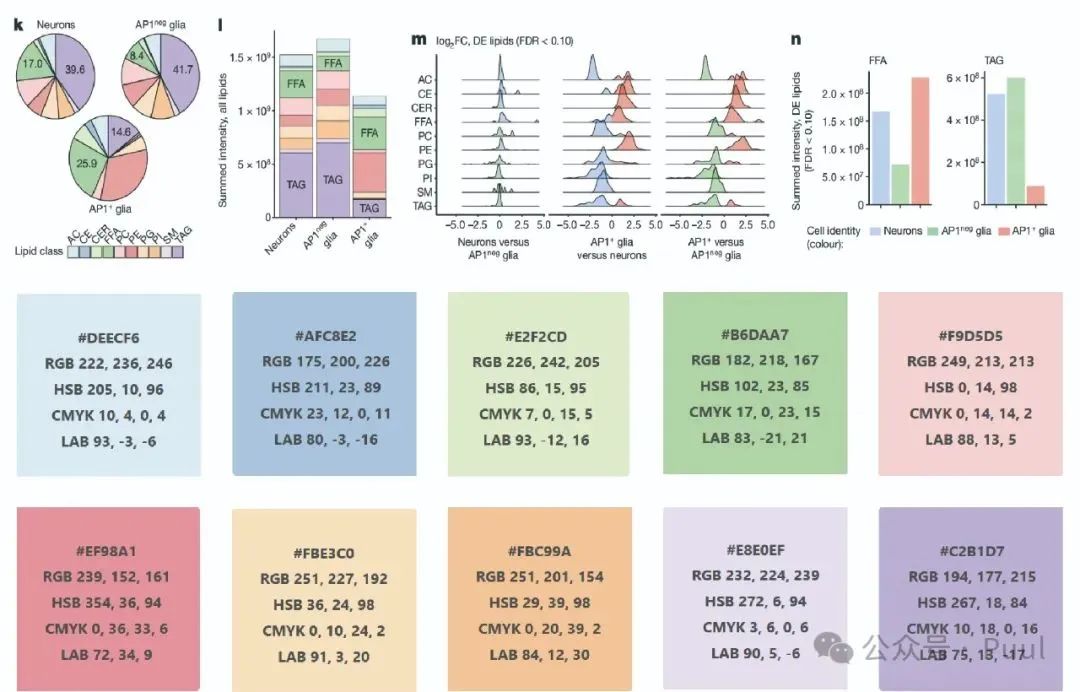
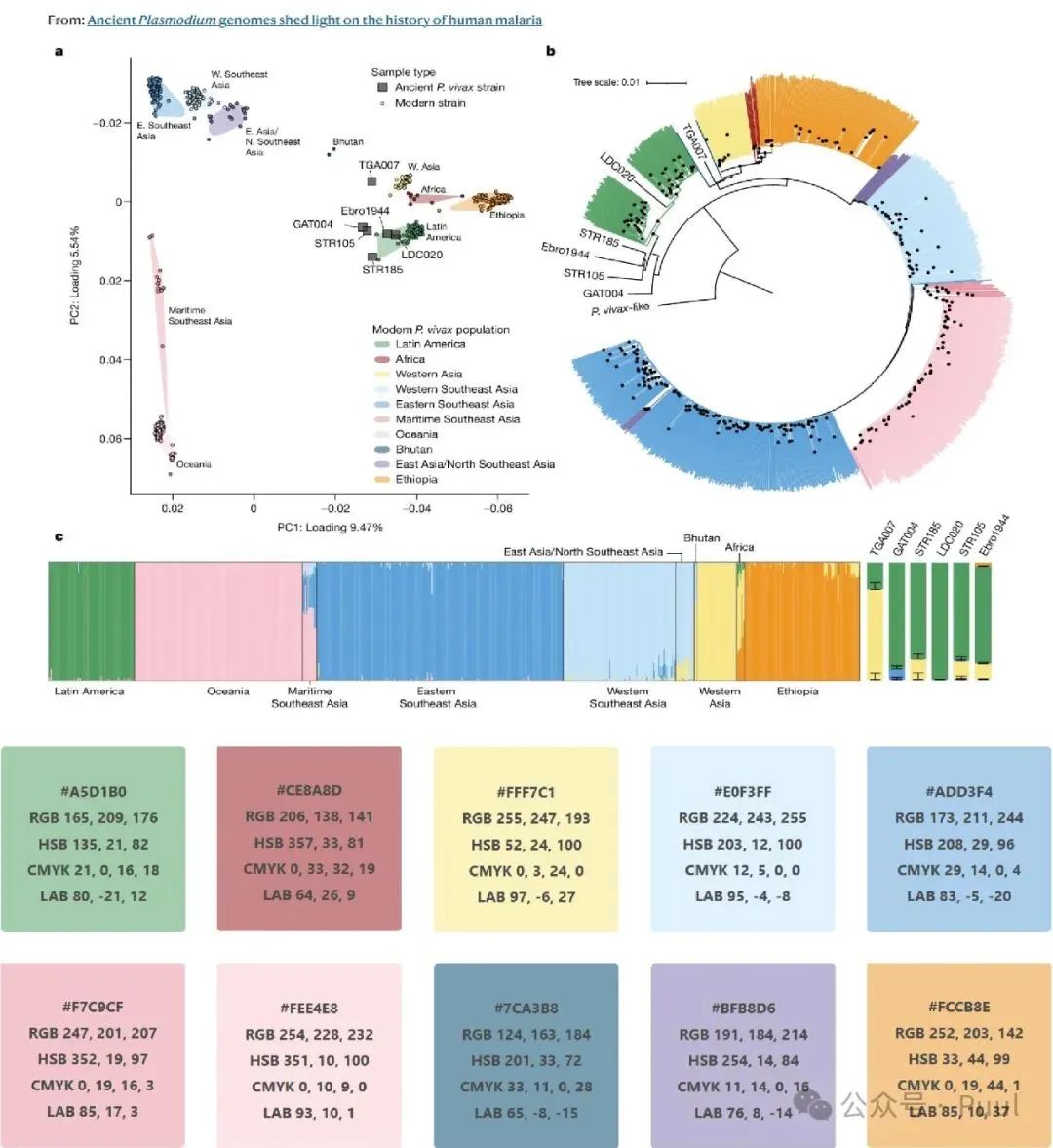
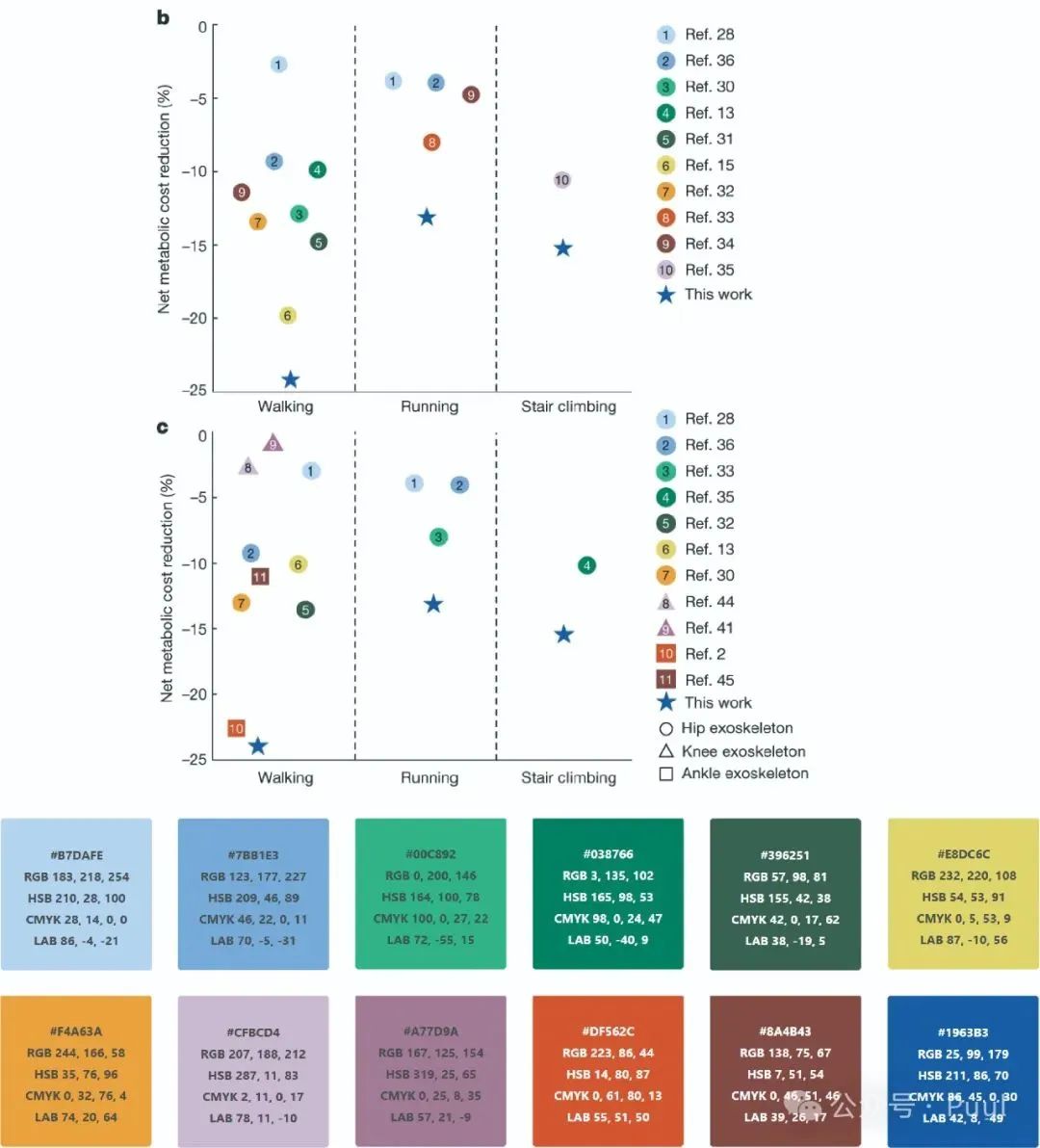
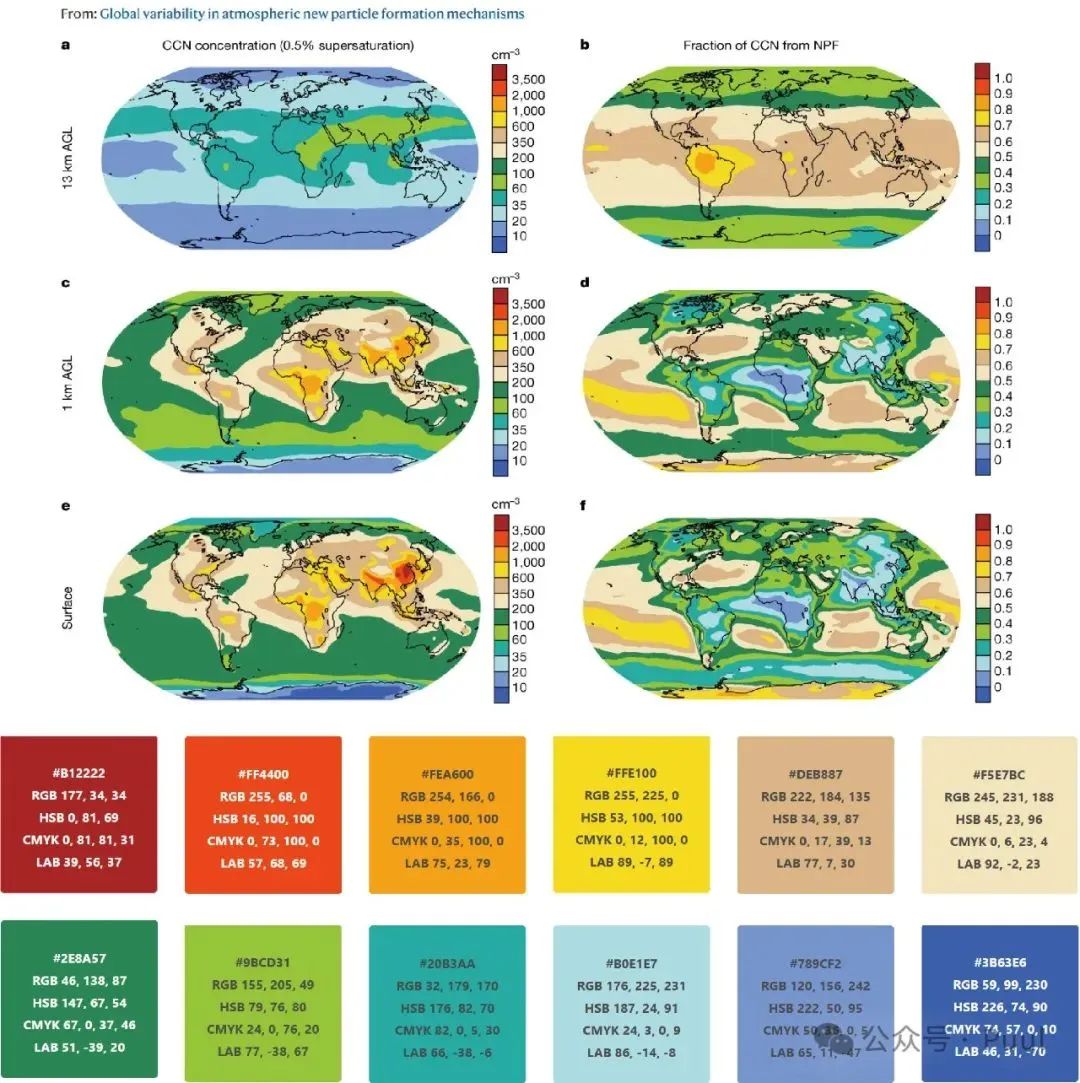
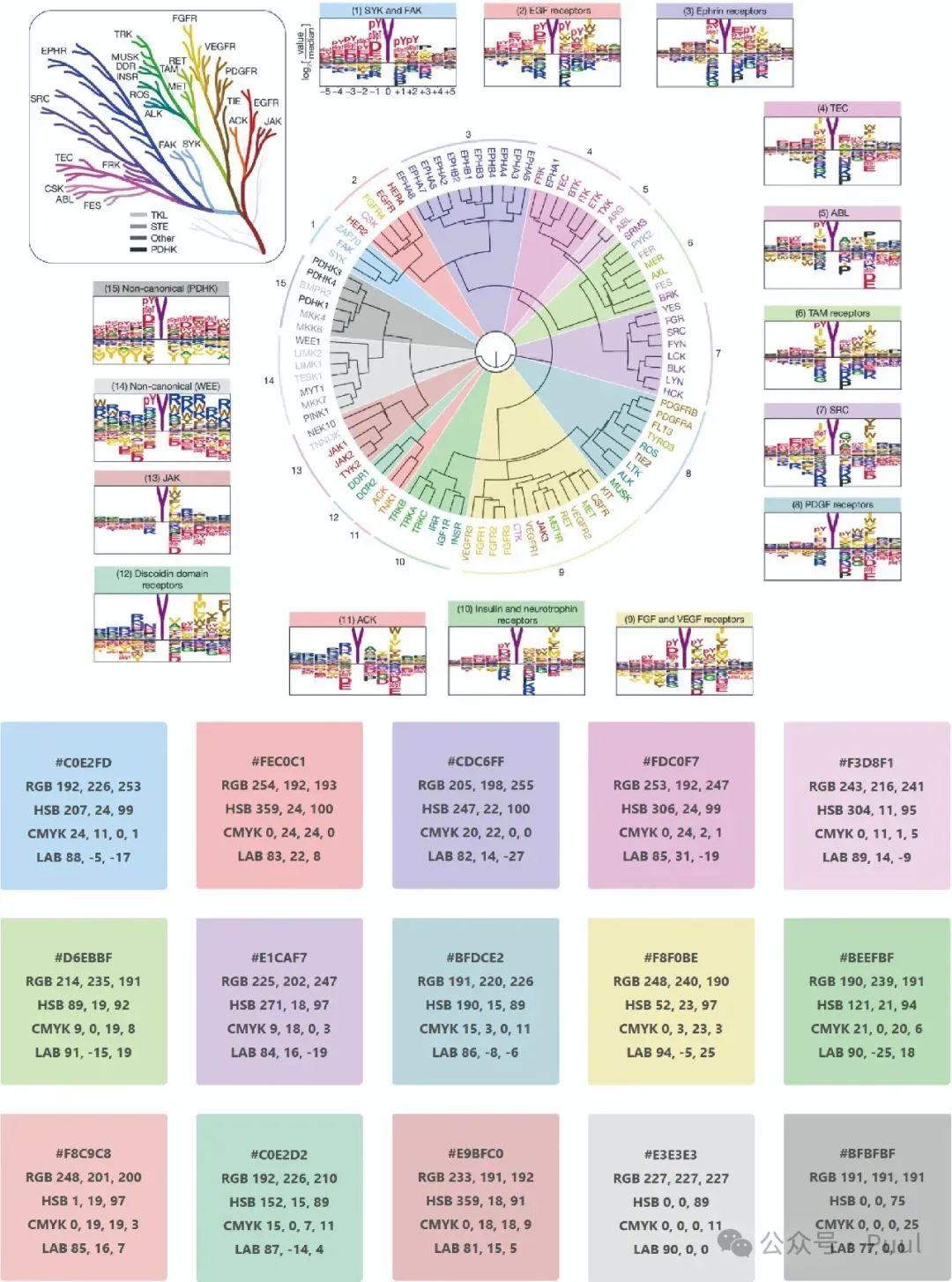
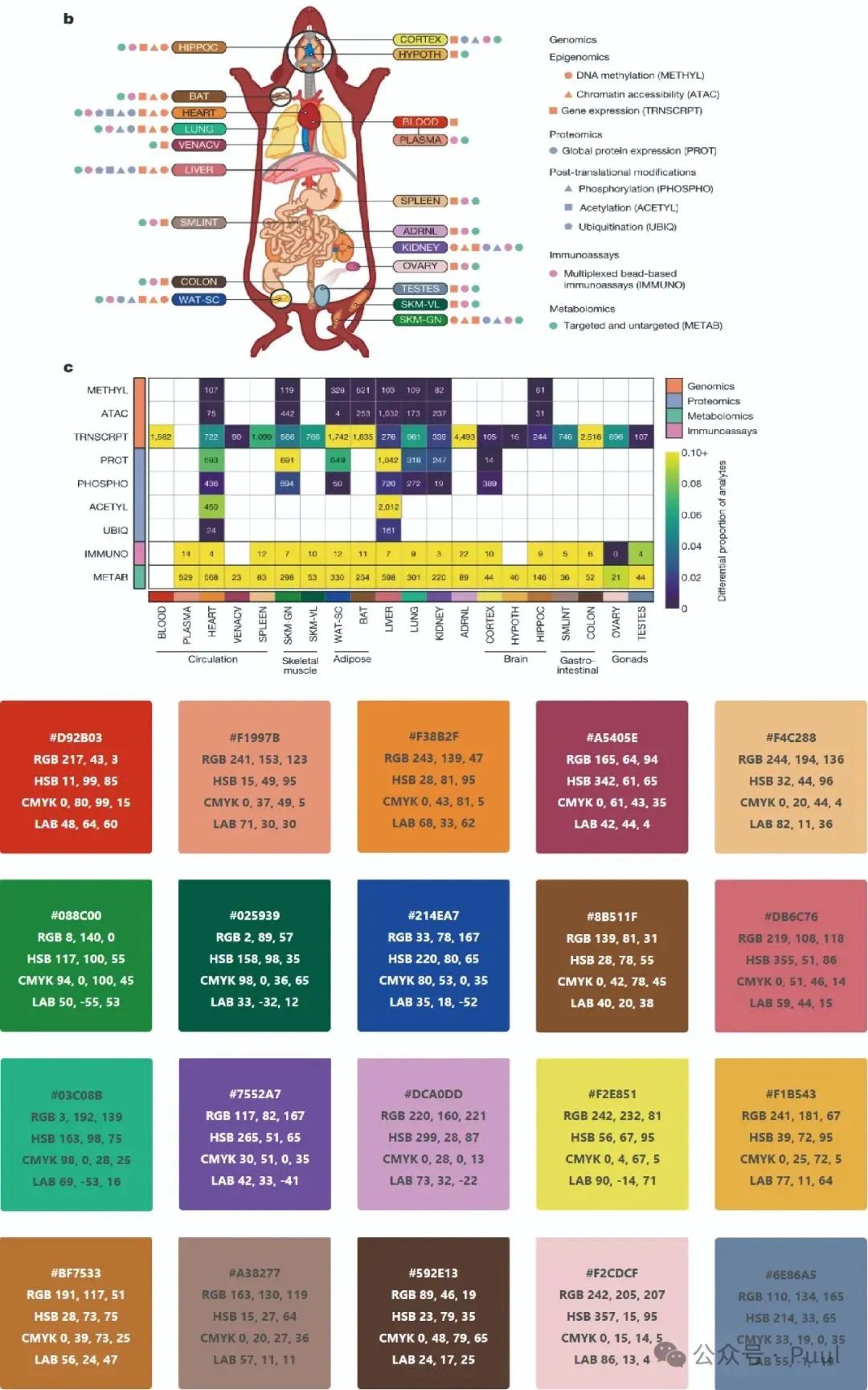
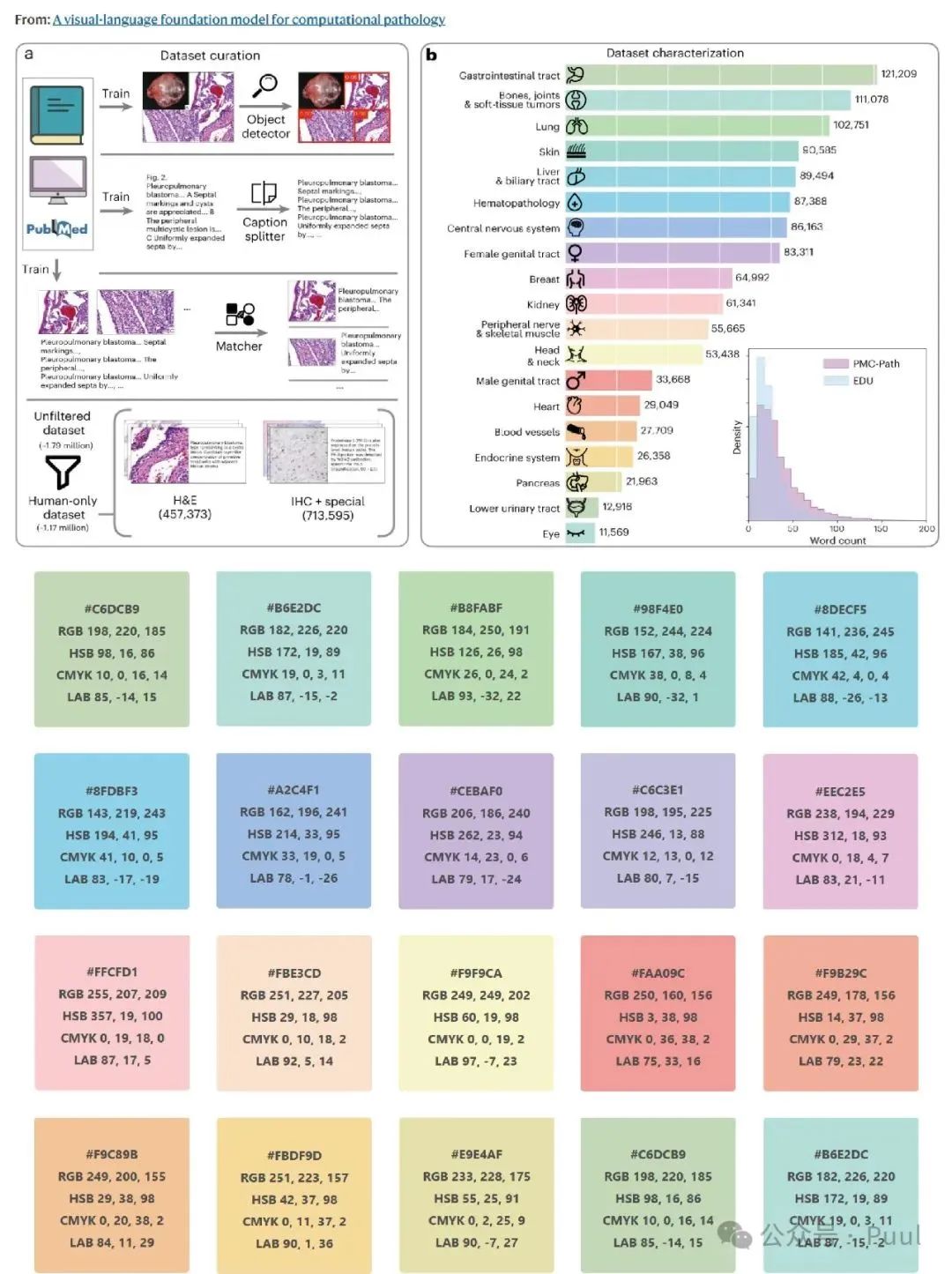
Descriptions and Models of Colors:
1. Hexadecimal: Hexadecimal color codes are a common method of color representation in web design and programming.
Representation: A combination of six digits and letters, with each pair representing a color channel (Red, Green, Blue).
Format: “#RRGGBB”, where “RR”, “GG”, and “BB” represent the values of red, green, and blue, ranging from 00 to FF (hexadecimal), which is 0 to 255 (decimal).
Example: “#FFFFFF” represents white, “#000000” represents black, “#FF0000” represents red.
2. RGB (Red, Green, Blue): The RGB color model is a method of color representation based on the additive principle of light, widely used in monitors, televisions, and photographic devices.
Representation: Colors are represented by mixing red (Red), green (Green), and blue (Blue) light.
Format: “RGB(R, G, B)”, with each value ranging from 0 to 255.
Example: “RGB(255, 0, 0)” represents red, “RGB(0, 255, 0)” represents green, “RGB(0, 0, 255)” represents blue, “RGB(255, 255, 255)” represents white.
3. HSB (Hue, Saturation, Brightness): The HSB color model (also known as HSV) aligns more closely with human perception of color.
Hue: Represents the basic attribute of color, ranging from 0 to 360 degrees. For example, 0 degrees is red, 120 degrees is green, and 240 degrees is blue.
Saturation: Represents the purity or intensity of color, ranging from 0% to 100%. 0% is gray, and 100% is the purest color.
Brightness: Represents the brightness level of color, ranging from 0% to 100%. 0% is black, and 100% is the brightest color.
Format: “HSB(H, S, B)”.
Example: “HSB(0, 100%, 100%)” represents red, “HSB(120, 100%, 100%)” represents green, “HSB(240, 100%, 100%)” represents blue.
4. CMYK (Cyan, Magenta, Yellow, Black): The CMYK color model is primarily used in printing, creating colors through subtractive mixing.
Representation: Colors are produced by mixing cyan (Cyan), magenta (Magenta), yellow (Yellow), and black (Key/Black).
Format: “CMYK(C, M, Y, K)”, with each value ranging from 0% to 100%.
Example: “CMYK(0%, 100%, 100%, 0%)” represents red, “CMYK(100%, 0%, 100%, 0%)” represents green, “CMYK(100%, 100%, 0%, 0%)” represents blue.
5. LAB (CIELAB): The LAB color model is based on human visual perception, aiming to represent colors more uniformly.
L: Represents lightness, ranging from 0 to 100. 0 represents black, and 100 represents white.
A: Represents the range from green to red, with positive values indicating red and negative values indicating green.
B: Represents the range from blue to yellow, with positive values indicating yellow and negative values indicating blue.
Format: “LAB(L, A, B)”.
Example: “LAB(53.24, 80.09, 67.20)”.
References:Amontree, Jacob, Xingzhou Yan, Christopher S. DiMarco, Pierre L. Levesque, Tehseen Adel, Jordan Pack, Madisen Holbrook, et al. 2024. “Reproducible Graphene Synthesis by Oxygen-Free Chemical Vapour Deposition.” Nature, May. https://doi.org/10.1038/s41586-024-07454-5.Argaw-Denboba, Ayele, Thomas S. B. Schmidt, Monica Di Giacomo, Bobby Ranjan, Saravanan Devendran, Eleonora Mastrorilli, Catrin T. Lloyd, et al. 2024. “Paternal Microbiome Perturbations Impact Offspring Fitness.” Nature 629 (8012): 652–59. https://doi.org/10.1038/s41586-024-07336-w.Chaudun, Fabrice, Laurena Python, Yu Liu, Agnes Hiver, Jennifer Cand, Brigitte L. Kieffer, Emmanuel Valjent, and Christian Lüscher. 2024. “Distinct Μ-Opioid Ensembles Trigger Positive and Negative Fentanyl Reinforcement.” Nature630 (8015): 141–48. https://doi.org/10.1038/s41586-024-07440-x.Chen, Xiaoyu, Fikri Birey, Min-Yin Li, Omer Revah, Rebecca Levy, Mayuri Vijay Thete, Noah Reis, et al. 2024. “Antisense Oligonucleotide Therapeutic Approach for Timothy Syndrome.” Nature 628 (8009): 818–25. https://doi.org/10.1038/s41586-024-07310-6.Desautels, Thomas A., Kathryn T. Arrildt, Adam T. Zemla, Edmond Y. Lau, Fangqiang Zhu, Dante Ricci, Stephanie Cronin, et al. 2024. “Computationally Restoring the Potency of a Clinical Antibody against Omicron.” Nature 629 (8013): 878–85. https://doi.org/10.1038/s41586-024-07385-1.Dong, Zehao, Mengwu Huo, Jie Li, Jingyuan Li, Pengcheng Li, Hualei Sun, Lin Gu, et al. 2024. “Visualization of Oxygen Vacancies and Self-Doped Ligand Holes in La3Ni2O7−δ.” Nature, June. https://doi.org/10.1038/s41586-024-07482-1.Dowding, Ian, and Christopher A. Schuh. 2024. “Metals Strengthen with Increasing Temperature at Extreme Strain Rates.” Nature 630 (8015): 91–95. https://doi.org/10.1038/s41586-024-07420-1.Dunant, Cyrille F., Shiju Joseph, Rohit Prajapati, and Julian M. Allwood. 2024. “Electric Recycling of Portland Cement at Scale.” Nature 629 (8014): 1055–61. https://doi.org/10.1038/s41586-024-07338-8.Guo, S.-A., Y.-K. Wu, J. Ye, L. Zhang, W.-Q. Lian, R. Yao, Y. Wang, et al. 2024. “A Site-Resolved Two-Dimensional Quantum Simulator with Hundreds of Trapped Ions.”Nature, May. https://doi.org/10.1038/s41586-024-07459-0.Han, Juntai, Ziwei Liu, Ross Woods, Tim R. McVicar, Dawen Yang, Taihua Wang, Ying Hou, Yuhan Guo, Changming Li, and Yuting Yang. 2024. “Streamflow Seasonality in a Snow-Dwindling World.” Nature 629 (8014): 1075–81. https://doi.org/10.1038/s41586-024-07299-y.Jin, Wenlong, Chi-Yuan Yang, Riccardo Pau, Qingqing Wang, Eelco K. Tekelenburg, Han-Yan Wu, Ziang Wu, et al. 2024. “Photocatalytic Doping of Organic Semiconductors.” Nature 630 (8015): 96–101. https://doi.org/10.1038/s41586-024-07400-5.Kim, Seongsoo, Ilya Svetlizky, David A. Weitz, and Frans Spaepen. 2024. “Work Hardening in Colloidal Crystals.” Nature, May. https://doi.org/10.1038/s41586-024-07453-6.Liu, Mei Hong, Benjamin M. Costa, Emilia C. Bianchini, Una Choi, Rachel C. Bandler, Emilie Lassen, Marta Grońska-Pęski, et al. 2024. “DNA Mismatch and Damage Patterns Revealed by Single-Molecule Sequencing.” Nature, June. https://doi.org/10.1038/s41586-024-07532-8.Luo, Qingyu, Evangeline G. Raulston, Miguel A. Prado, Xiaowei Wu, Kira Gritsman, Karley S. Whalen, Kezhi Yan, et al. 2024. “Targetable Leukaemia Dependency on Noncanonical PI3Kγ Signalling.” Nature 630 (8015): 198–205. https://doi.org/10.1038/s41586-024-07410-3.Luo, Shuzhen, Menghan Jiang, Sainan Zhang, Junxi Zhu, Shuangyue Yu, Israel Dominguez Silva, Tian Wang, et al. 2024. “Experiment-Free Exoskeleton Assistance via Learning in Simulation.” Nature 630 (8016): 353–59. https://doi.org/10.1038/s41586-024-07382-4.Ma, Shoucai, Chunpeng An, Aaron W. Lawson, Yu Cao, Yue Sun, Eddie Yong Jun Tan, Jinheng Pan, et al. 2024. “Oligomerization-Mediated Autoinhibition and Cofactor Binding of a Plant NLR.” Nature, June. https://doi.org/10.1038/s41586-024-07668-7.Mahon, Michael B., Alexandra Sack, O. Alejandro Aleuy, Carly Barbera, Ethan Brown, Heather Buelow, David J. Civitello, et al. 2024. “A Meta-Analysis on Global Change Drivers and the Risk of Infectious Disease.” Nature 629 (8013): 830–36. https://doi.org/10.1038/s41586-024-07380-6.Michel, Megan, Eirini Skourtanioti, Federica Pierini, Evelyn K. Guevara, Angela Mötsch, Arthur Kocher, Rodrigo Barquera, et al. 2024. “Ancient Plasmodium Genomes Shed Light on the History of Human Malaria.” Nature, June. https://doi.org/10.1038/s41586-024-07546-2.Miyamoto, Yu, Junichi Kikuta, Takahiro Matsui, Tetsuo Hasegawa, Kentaro Fujii, Daisuke Okuzaki, Yu-chen Liu, et al. 2024. “Periportal Macrophages Protect against Commensal-Driven Liver Inflammation.” Nature 629 (8013): 901–9. https://doi.org/10.1038/s41586-024-07372-6.MoTrPAC Study Group, Primary authors, Lead Analysts, David Amar, Nicole R. Gay, Pierre M. Jean-Beltran, Lead Data Generators, et al. 2024. “Temporal Dynamics of the Multi-Omic Response to Endurance Exercise Training.” Nature 629 (8010): 174–83. https://doi.org/10.1038/s41586-023-06877-w.Needham, Lisa-Maria, Carlos Saavedra, Julia K. Rasch, Daniel Sole-Barber, Beau S. Schweitzer, Alex J. Fairhall, Cecilia H. Vollbrecht, et al. 2024. “Label-Free Detection and Profiling of Individual Solution-Phase Molecules.” Nature629 (8014): 1062–68. https://doi.org/10.1038/s41586-024-07370-8.Neupane, Sujaya, Ila Fiete, and Mehrdad Jazayeri. 2024. “Mental Navigation in the Primate Entorhinal Cortex.” Nature, June. https://doi.org/10.1038/s41586-024-07557-z.Niepoth, Natalie, Jennifer R. Merritt, Michelle Uminski, Emily Lei, Victoria S. Esquibies, Ina B. Bando, Kimberly Hernandez, et al. 2024. “Evolution of a Novel Adrenal Cell Type That Promotes Parental Care.” Nature 629 (8014): 1082–90. https://doi.org/10.1038/s41586-024-07423-y.Reimink, Jesse R., and Andrew J. Smye. 2024. “Subaerial Weathering Drove Stabilization of Continents.” Nature 629 (8012): 609–15. https://doi.org/10.1038/s41586-024-07307-1.Ri, Keiken, Tsai-Hsuan Weng, Ainara Claveras Cabezudo, Wiebke Jösting, Yu Zhang, Andre Bazzone, Nancy C. P. Leong, et al. 2024. “Molecular Mechanism of Choline and Ethanolamine Transport in Humans.” Nature 630 (8016): 501–8. https://doi.org/10.1038/s41586-024-07444-7.Riris, Philip, Fabio Silva, Enrico Crema, Alessio Palmisano, Erick Robinson, Peter E. Siegel, Jennifer C. French, et al. 2024. “Frequent Disturbances Enhanced the Resilience of Past Human Populations.” Nature 629 (8013): 837–42. https://doi.org/10.1038/s41586-024-07354-8.Wan, Jun-Nan, Sheng-Wei Wang, Andrew R. Leitch, Ilia J. Leitch, Jian-Bo Jian, Zhang-Yan Wu, Hai-Ping Xin, et al. 2024. “The Rise of Baobab Trees in Madagascar.” Nature 629 (8014): 1091–99. https://doi.org/10.1038/s41586-024-07447-4.Source: Pu.Special Statement: All works indicated as sourced from this public account are reprinted from other media, and the copyright belongs to the original authors. This WeChat reprint is for non-commercial educational and research purposes. If there are any copyright issues with the reprinted materials, please contact us immediately, and we will make changes or delete the relevant articles to ensure your rights. For reprints, please contact the original source. (Some images are sourced from the internet; if there is any infringement, please contact us for deletion.)
Thank you for taking the time to read this article.

.

.

Come read this article.
Editor: Yao HuiProofreader: Liang FujunReviewer: Zhang Qiang

Long press the image, scan the QR code, and follow us!

Chinese Surface EngineeringChinese, Core Journal of Science and Technology
Indexed in ESCI, EI, Scopus, etc.Article recommendations / Manuscript inquiries / Journal browsing
