In a previous article, we discussed the prediction of CpG islands. Once a CpG island is identified, it is necessary to detect the methylation status of the corresponding region. There are various methods to detect methylation status, but the currently recognized gold standard is the bisulfite sequencing PCR (BSP).
BSP Principle
Genomic DNA is treated with sodium bisulfite, where all unmethylated cytosines (C) are converted to uracils (U), while methylated cytosines remain unchanged.
After bisulfite treatment of genomic DNA, BSP primers are designed to amplify the target fragment. At this point, all uracils (U) are converted to thymines (T), and sequencing the PCR products allows us to determine whether the CpG sites are methylated.
BSP Experimental Process:
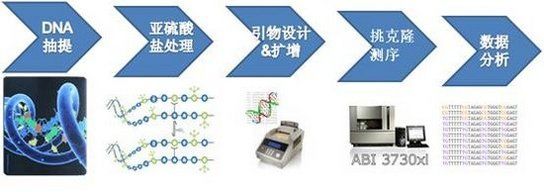
Both DNA extraction and bisulfite treatment can refer to the corresponding reagent kits for operation.
The primer design website is as follows:http://www.urogene.org/cgi-bin/methprimer2/MethPrimer.cgi
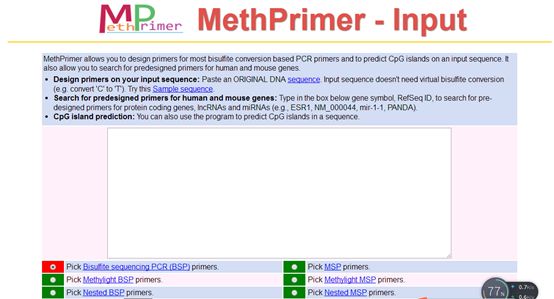
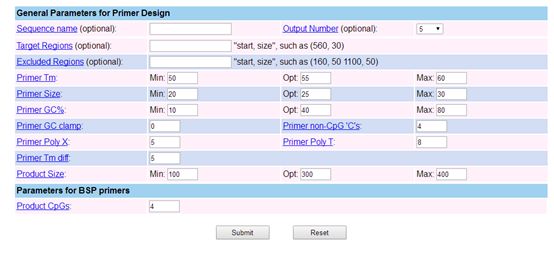
The website’s primer design interface is shown in the above image, where you can choose what type of primer to design, here selecting to design BSP primers.
This website can also provide functions such as CpG island prediction, so you can select the option for primer design after predicting CpG islands, as shown in the following image:
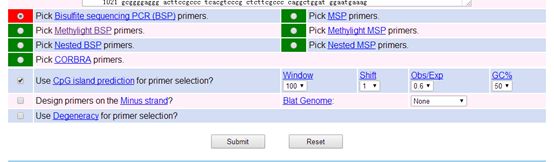
Other parameter selections can remain at their default values, still taking the human GAPDH promoter sequence as an example.
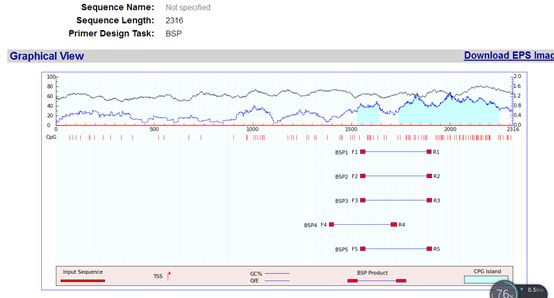
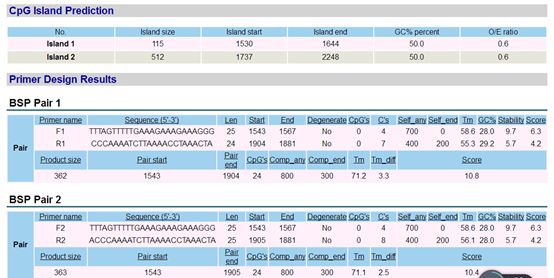
The design results are shown above, predicting two CpG islands. The designed primers based on the measured results are shown in the above image.
Additionally, you can adjust the relevant parameters for primer design according to your needs, as shown in the following image:
The prediction results also provide information on the changes in sequences before and after bisulfite treatment, the positions of the primers, and the positions of the CpG sites:


There are also related precautions in primer design:
The DNA used for BSP experiments is treated with bisulfite, converting unmethylated C bases to U bases, while methylated C bases remain unchanged. This transforms the differences in methylation into differences in bases, so when designing primers for PCR amplification, it is important to note:
1. The primer design must not include CpG sites to avoid differences between methylated and unmethylated DNA.
2. The amplified fragment should include as many CpG sites as possible. The BSP amplification fragment length is 300-500bp, and the more CpG sites included, the higher the score of the primers.
After completing the BSP primer design, PCR amplification should be performed for direct sequencing or TA cloning sequencing analysis of methylation frequency.
BSP Direct Sequencing Results
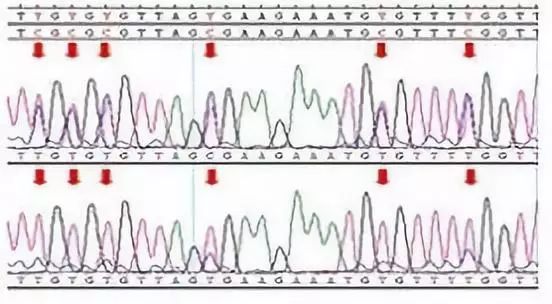
BSP Dot Plot

Empty points represent unmethylated CpG sites, while filled points represent methylated CpG sites.
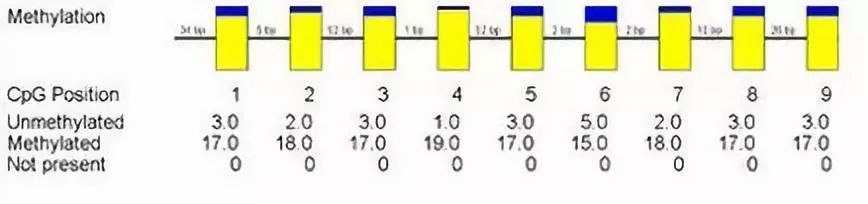
BSP Bar Chart
Scan to follow
