Welcome to the Live Broadcast on Bioinformatics Home:
Integration Analysis of Metagenomics (16S and Metagenome) and Metabolomics
Broadcast starts on July 9 at 20:30, scan the QR code below to register for free

Abstract
The oncogene fusion gene SPI-1 frequently appears in cases of T-cell acute lymphoblastic leukemia (T-ALL), but it is not sufficient to drive leukemia development. Here, the authors used a conditional bone marrow transplantation mouse model to show that the SPI1 fusion combined with activated NRAS mutations drives an immature T-ALL in vivo. The addition of the oncogenic fusion on the basis of NRAS mutations also leads to an increased frequency of leukemia stem cells. Mechanistically, the oncogenic activity of the fusion can be eliminated by knocking out the β-catenin binding domain within transcription factor 7-SPI1 or by using antagonists that inhibit TCF/β-catenin interactions. Both drug and gene suppression lead to downregulation of SPI1 targets. In conclusion, the authors’ results suggest that TCF7-SPI1 leukemia is susceptible to targeted therapy against TCF/β-catenin interactions.
Paper ID
Title:Oncogenic cooperation between TCF7-SPI1 and NRAS(G12D) requires β-catenin activity to drive T-cell acute lymphoblastic leukemia
Translated Title:TCF7-SPI1 and NRAS(G12D) oncogenic cooperation requires β-catenin activation to drive T-cell acute lymphoblastic leukemia
Journal:Nature Communications
Impact Factor:14.914
Publication Date:July 6, 2021
Corresponding Author Affiliation:University of New South Wales, Australia
DOI: https://doi.org/10.1038/s41467-021-24442-9
Main Content
Identification of T-ALL cases positive for TCF7-SPI1 and NRAS mutations
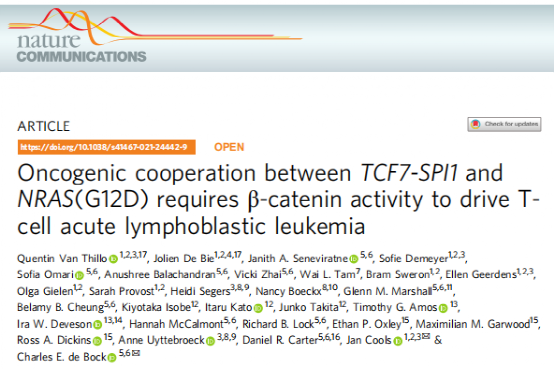
The peripheral blood cells are from a newly diagnosed 6-year-old boy with immature CD4/CD8 double-negative T-ALL, who underwent whole-genome and RNA sequencing, revealing translocations between chromosomes 5 and 11 and coexisting mutations of NRAS. Concurrent mutations were found in genes such as NOTCH1, Krüppel-like factor 9, Cyclin-dependent kinase inhibitor 2A, Sidekick cell adhesion molecule 1, T-cell receptor gamma, and transducin-like 1 X-linked receptor 1. The translocation breakpoints occurred in intron 4-5 of TCF7 and intron 2-3 of SPI1, generating a predicted fusion transcript linking exons 1-4 of TCF7 and exons 3-5 of SPI1, which was validated through long-read nanopore and Sanger sequencing. At the protein level, the predicted fusion product aggregates the first 182 amino acids of TCF1, including the N-terminal β-catenin binding region, and the terminal amino acids 52 to 271 of PU.1, forming a protein of 402 amino acids that still retains the DNA binding domain region of PU.1.
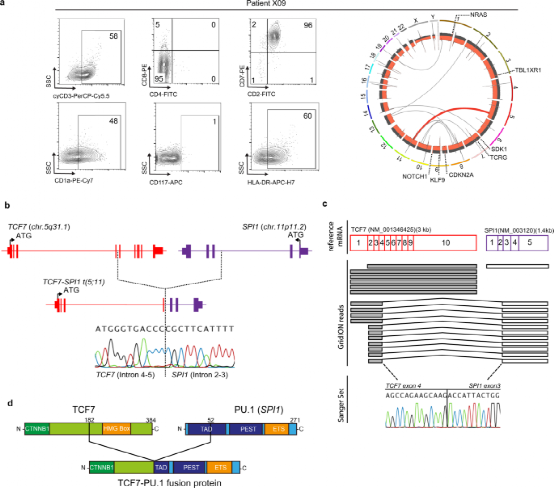
Figure 1: Identification and characterization of TCF7-SPI1 fusion in T-cell acute lymphoblastic leukemia patient X09.
Single-cell RNA sequencing revealed that high expression of SPI1 is associated with NRAS, stem cells, and Wnt/β-catenin signaling cell characteristics..
Single-cell RNA sequencing was performed on six primary T-ALL samples, including patient X09, to determine the transcriptional determinants of this T-ALL subtype. Clustering of a total of 13,848 cells from the aggregated samples identified 22 cell clusters representing malignant T cells or normal cells, including NK-T cells, B cells, monocytes, and erythrocytes. Focusing only on 11,620 malignant T cells, the patients with TCF7-SPI1 fusion were distinct from other T-ALL samples, showing significantly higher SPI1 expression. Gene expression analysis also showed that in fusion-positive cases, SPI1 expression was associated with high levels of hematopoietic stem cell markers such as CD34 and low levels of mature T cell markers such as CD3D and CD1E, consistent with the immature phenotype observed in flow cytometry analysis.
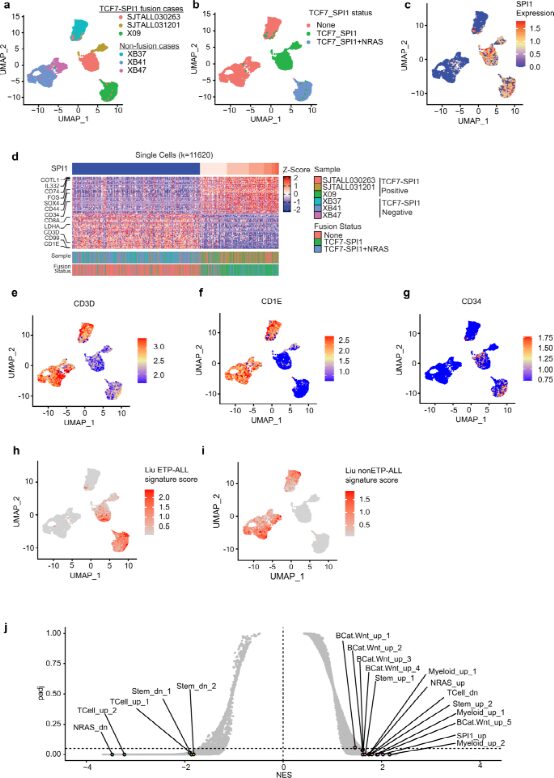
Figure 2: Single-cell RNA sequencing revealed that high expression of SPI1 is associated with NRAS, stem cells, and Wnt/β-catenin signaling cell characteristics.
The authors next attempted to determine the transcriptional comparison between fusion-positive cases and other defined T-ALL subtypes. They found that fusion cases clustered with early T cell precursor cases and only immature Lim domain 2/lymphoblastic leukemia-associated hematopoietic regulators 1 cases from the dataset of Liu et al., suggesting that although fusion cases have an immune phenotype, their transcriptome is similar to ETP-ALL. Therefore, using published transcriptome data, the authors subsequently generated gene features from ETP-ALL and non-ETP-ALL patients. Using single-cell RNA sequencing data from normally developing human thymocytes, the features generated by Liu and Zhang were validated, confirming the highest enrichment in designated ETP regions. In contrast, Liu’s non-ETP-ALL and Zhang’s T-ALL features were most enriched in designated double-positive and αβT-cell entry regions. Applying these features to the authors’ data, the three TCF7-SPI1 fusion-positive cases showed a positive enrichment for ETP-ALL features and a negative enrichment for complementary T-ALL features.
Combining the extensive RNA sequencing data of these six patients with the cohort described by Seki et al., it was further found that patients carrying both TCF7-SPI1 fusion and NRAS mutations had higher ETP-ALL features compared to those with only TCF7-SPI1 fusion. The authors also used GSEA analysis to determine the signaling pathways associated with SPI1 expression, finding positive enrichment of bone marrow, stem, and Wnt/β-catenin gene features. Overall, these data highlight that the fusion-positive transcriptome is enriched for ETP features and there is a strong link between TCF7-SPI1 fusion status, RAS activation, and the Wnt/β-catenin pathway.
TCF7-SPI1 requires mutated NRAS to induce in vitro transformation
Given the high frequency of co-occurring mutations with NRAS and its association with NRAS signaling, the authors sought to investigate whether TCF7-SPI1 fusion cooperates with oncogenic NRAS to drive T-ALL. They generated two sets of retroviral vectors that express TCF7-SPI1 and/or NRAS(G12D) in a constitutive or inducible manner using the viral P2A sequence. Immunoblotting confirmed that both TCF7-SPI1 fusion and/or NRAS could effectively constitute or induce expression when transduced into Ba/F3 cells and Ba/F3-Cre cells. The transformation potential was first determined in Ba/F3 cells expressing either NRAS(G12D) alone or wild-type and Cre-positive cells combined with TCF7-SPI1. Only in the presence of NRAS(G12D) could the cells grow in an interleukin-3-independent manner, indicating that NRAS(G12D) protein provides necessary proliferation and survival signals to lymphocytes. Combining the authors’ inducible vector system with a physiologically relevant Cre-positive precursor T-cell culture model, all transduced cells grew in the presence of Delta-like ligand 4, interleukin-7, and stem cell factor. Notably, the induced expression of NRAS(G12D) +/- TCF7-SPI1 provided growth and survival signals that could transform cells to Scf-, and to a lesser extent to Il7-, but both still relied on Notch signaling through Dll4. Therefore, these results suggest that NRAS(G12D) alone is sufficient and necessary for the transition of these two in vitro cell systems to cytokine-independent growth.
Figure 3: The presence of NRAS(G12D) mutation is a necessary condition for transformation to cytokine-independent growth in vitro.
The expression of TCF7-SPI1 and NRAS(G12D) leads to aggressive T-cell leukemia in a mouse bone marrow transplantation model.
The authors next attempted to determine whether the combination of TCF7-SPI1 and NRAS synergistically drives T-ALL in vivo. For this purpose, hematopoietic stem and progenitor cells (HSPCs) isolated from the bone marrow of CD2-Cre positive C57BL/6J mice were transduced with different lox66/71 vectors to restrict the expression of TCF7-SPI1 and/or NRAS in developing T cells. Wild-type recipient mice receiving only TCF7-SPI1 transduced cells did not develop leukemia during the 200-day observation period post-injection. Recipient mice receiving TCF7-SPI1 + NRAS(G12D) developed leukemia, with a latency similar to that of mice receiving NRAS(G12D) alone. Both were confirmed as acute leukemia, and secondary subjects succumbed to leukemia with significantly reduced latency.
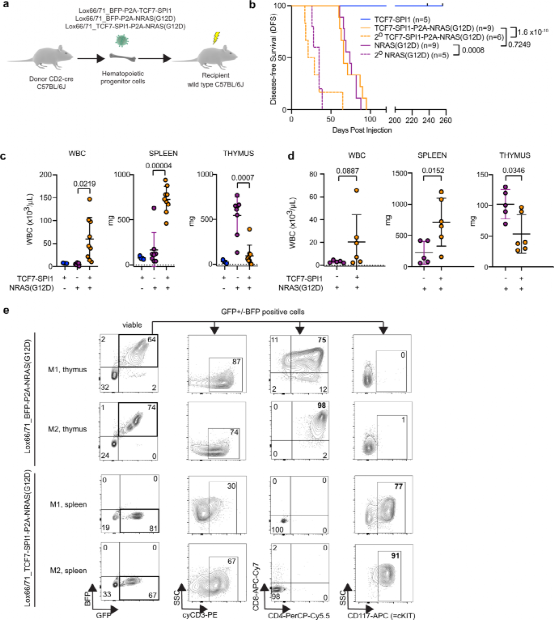
Figure 4: Conditional co-expression of TCF7-SPI1 fusion and NRAS(G12D) in developing T cells synergistically produces immature T-cell acute lymphoblastic leukemia.
Although NRAS(G12D) alone and TCF7-SPI1-P2A-NRAS(G12D) both induce leukemia with similar latency, there are significant differences in disease manifestation and immune phenotype. Mice expressing NRAS(G12D) alone in developing T cells showed thymic enlargement but little increase in white blood cell count or leukemic infiltration of the spleen. In contrast, mice co-expressing NRAS(G12D) and TCF7-SPI1 exhibited leukocytosis and splenomegaly but no thymic enlargement. This phenotype remained unchanged after secondary transplantation. Further phenotyping of the resulting leukemias by flow cytometry revealed that NRAS(G12D) leukemia cells exhibited a more differentiated CD4+/CD8+ immune phenotype, while TCF7-SPI1+NRAS(G12D) leukemia cells displayed an immature immune phenotype of CD4-/CD8-, CD3+, and CD117+. The authors then performed in vivo limiting dilution assays to assess the frequency of leukemia stem cells, indicating a significantly higher LSC frequency in TCF7-SPI1-P2A-NRAS(G12D) mice compared to NRAS(G12D) alone.
TCF7-SPI1-induced immature leukemia requires upstream β-catenin activity
Given the Wnt/β-catenin signaling features associated with SPI1 expression found in single-cell data and the presence of a β-catenin binding domain in the N-terminal region of the fusion protein, the authors next sought to determine whether the N-terminal β-catenin binding domain of TCF1 within the fusion protein is also important in leukemia development. It is well known that TCF proteins act as typical Wnt signaling mediators alongside β-catenin. Furthermore, β-catenin has previously been recognized as critical for LSC self-renewal and has recently been linked to maintaining LSC stemness and SPI1 expression. Therefore, the authors established a new bone marrow transplantation model, selectively removing the first 55 amino acids of TCF1 that constitute the β-catenin binding site. Similar to the leukemia occurring in NRAS(G12D) mice, recipient mice receiving only ΔβCat-TCF7-SPI1 exhibited thymic enlargement, limited splenic infiltration, a more mature CD4+/CD8+ immune phenotype, and similar disease latency. This indicates that the loss of the β-catenin binding domain cancels the effect of the fusion, leading to disease driven only by NRAS.
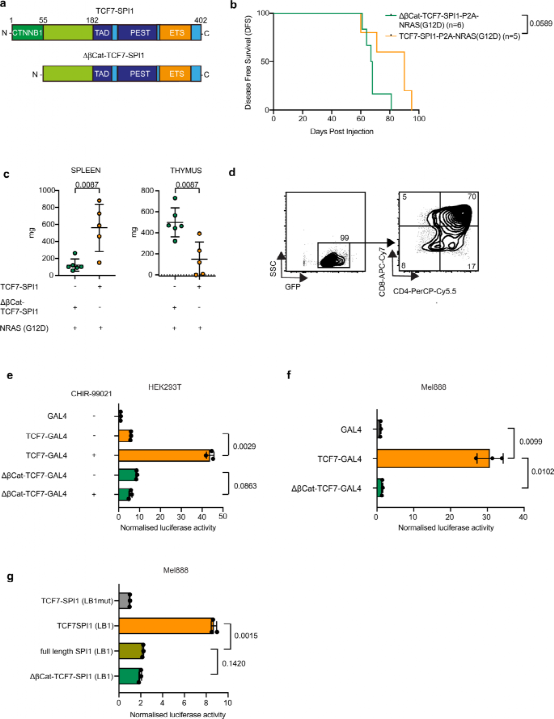
Figure 5: The N-terminal β-catenin binding site of TCF1-PU.1 fusion protein is essential for in vivo transcriptional activity and the development of ETP-ALL.
To further determine whether the absence of β-catenin binding is the reason for altered disease manifestation, the authors constructed a series of GAL4 DNA binding domain fusion constructs containing either the N-terminal TCF7 portion of TCF7-SPI1 fusion or the truncated ΔβCat-TCF7. In the presence or absence of the GSK3β inhibitor CHIR-99021 to activate upstream Wnt signaling and increase β-catenin levels, luciferase reporter gene expression assays were performed to assess transcriptional activity in HEK293T cells. Activation of Wnt signaling only resulted in increased luciferase activity for the full-length construct, while the ΔβCat-TCF7 construct did not, which was characteristically observed in the melanoma Mel888 cell line due to the S37F mutation, which provides constitutive activity to β-catenin. The authors observed similar differences between TCF7-SPI1 and ΔβCat-TCF7-SPI1 fusion constructs in luciferase reporter experiments using Lambda B1 or Fes promoters, both of which have endogenous SPI1 binding sites. Overall, SPI1-induced luciferase activity was lower than that induced by the fusion TCF7-SPI1, similar to the levels of ΔβCat-TCF7-SPI1 fusion. In summary, these data suggest that the immature T-ALL phenotype induced by TCF7-SPI1 fusion is partly driven by upstream β-catenin activity.
Genetic and small molecule antagonism of TCF7-SPI1 fusion leads to leukemia phenotype differentiation
To further determine the functional role of TCF7-SPI1 fusion in leukemia maintenance and survival, the authors employed inducible knockout using a Tet-On vector system to express short hairpin RNAs. At the pre-determined endpoint, the expression of the fusion was reduced by 50%, but there was no reduction in leukemia burden in peripheral blood, nor was there a decrease in spleen weight following doxycycline-induced knockout of the fusion. Shockingly, CD4 and CD8 significantly increased, while CD117 surface expression disappeared, along with changes at the mRNA level. This increase in phenotypic differentiation was further supported by RNA-seq analysis, showing a significant reduction in ETP-ALL characteristics after knockout of the fusion.
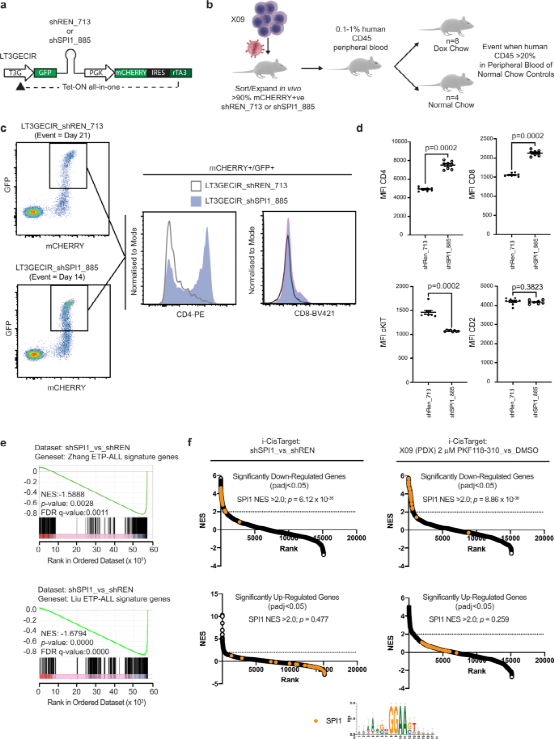
Figure 6: Genetic and small molecule antagonism of TCF7-SPI1 fusion leads to leukemia phenotype differentiation.
Using iCisTarget for global analysis of differentially expressed genes to identify common regulatory elements, it was found that significantly downregulated genes were highly enriched in SPI1 binding patterns. To confirm the requirement of β-catenin for the oncogenic function of TCF7-SPI1 and to verify the impact of fusion knockout on differentiation, the authors utilized the small molecule β-catenin/TCF antagonists PKF 118-310. PKF 118-310 completely eliminated luciferase signals generated by TCF7-GAL4 or TCF7-SPI1 in Mel888 cells, characterizing the gene removal of the β-catenin binding domain. Furthermore, similar to the iCisTarget results after fusion knockout, treatment with PKF-118-310 in three TCF7-SPI1 fusion-positive PDX samples also showed enrichment of SPI1 patterns in downregulated genes and reduced expression of cKIT. However, compared to fusion-negative T-ALL cases, in vitro treatment with PKF 118-310 did not show specific cytotoxicity for fusion-positive T-ALL cases.
In summary, these data suggest that the interaction between β-catenin and TCF1 is crucial for enhancing the oncogenic function of TCF7-SPI1 fusion, and its inhibition leads to downregulation of SPI1 targets and differentiation of leukemia cells.
Conclusion
In conclusion, the authors demonstrate that TCF7-SPI1 cooperates with NRAS to drive an aggressive immature T-cell leukemia in vivo, and it is possible to target this fusion protein by inhibiting the protein-protein interaction between TCF1 and β-catenin. Therefore, patients with TCF7-SPI1 fusion may benefit from improved pharmacological targeting of TCF/β-catenin interactions.
Original link: https://www.nature.com/articles/s41467-021-24442-9
Scan to reply with the number: 88
Receive an 88 yuan coupon for the replay of the live broadcast.

The Medical Plus platform has trained and guided thousands of students on SCI articles and National Natural Science Foundation applications, a considerable number of whom have published SCI articles or obtained National Natural Science Foundation projects.
The Medical Plus platform has hundreds of National Natural Science Foundation reviewers (professors with more than three National Natural Science Foundation hosting experiences, with a single SCI impact factor of over 10), all of whom have rich experience in reviewing SCI articles and National Natural Science Foundation applications.

[Medical Plus] Topic/article, expert precise matching, topic guidance design review consultation
Research topics in fields such as tumors, microbiomes and metabolism, epigenetic m6A, single-cell, multi-omics, non-coding RNA exosomes, traditional Chinese medicine, neurology, endocrinology, ophthalmology, orthopedics, cardiology, etc. are welcome to contact Medical Plus for review and guidance, we always have a peer expert who can precisely match with you, we call them “small peer experts”.

Expert consultation/class registration or card purchase, please scan to consult (invitation letters available, invoices can be issued)


The tumor experts at Medical Plus are providing one-on-one guidance for a tumor-related National Natural Science Foundation project application.
| 2021.7.3-4 | High-level Biomedical Topic Design and High-quality SCI Writing Intensive Class, Hebei Medical University First Hospital | Medical Plus Tencent Meeting |
| 2021.7.2-4 | TCGA, GEO Bioinformatics High-throughput Data Mining Two-day Intensive Class | Medical Plus Tencent Meeting |
| 2021.7.3-4 | RNA Methylation (M6A) Data Analysis and Topic Design Intensive Class | Medical Plus Tencent Meeting |
| 2021.7.24-25 | Single-cell Sequencing Data Analysis and Topic Design Online Intensive Class | Medical Plus Tencent Meeting |
| 2021.7.24-25 | Evidence-based Medicine Meta-analysis and Network Meta-analysis Intensive Class, Hebei Medical University First Hospital | Tencent Meeting |
| 2021.7.31-8.1 | Gut Microbiome and Metabolomics Topic Design Online Intensive Class | Medical Plus Tencent Meeting |
| 2021.7.31-8.1,8.2~5 | Whole Transcriptomics Training Camp | Medical Plus Tencent Meeting |
| 2021.7.31-8.1 | Research Strategies and Skills of Network Pharmacology of Traditional Chinese Medicine and Molecular Docking Online Intensive Class | Shanghai Zhongxing Hetai Hotel/Tencent Meeting |
| 2021.8 | National R Language Data Analysis Practical Technology Online Learning Class (Theory + Practice Class) | Shanghai Zhongxing Hetai Hotel/Tencent Meeting |
| 2021.8.14-15 | Clinical Prediction Models | Shanghai Zhongxing Hetai Hotel/Tencent Meeting |
| 2021.8 | Advanced Scientific Research Charting On-site Intensive Class, | Tencent Meeting |

Medical Plus Bioinformatics + Basic + Clinical Research Practical Training, taught by research experts,
teaching you how to fish, WeChat interactive Q&A, quickly improving research capabilities!
High-quality topics and articles with expert one-on-one guidance
To help new and old students of Medical Plus systematically master bioinformatics, basic research, and clinical research ideas and skills, thereby comprehensively, systematically, and rapidly enhancing students’ research innovation capabilities. Considering the current actual situation during the pandemic, we have set up more than ten online practical learning classes (opening monthly), to ensure the learning effect, we use the online meeting platform for teaching + video playback, with good interactive experience! Research units are welcome to contact us for appointments, and if needed, experts and professors from Medical Plus can also provide one-on-one topic guidance, teaching you how to fish!
[Bioinformatics Section—For detailed introduction, please click on the image below corresponding course title, register to first receive course videos and materials, you can sign up for a single course or buy a card to participate in the whole year’s learning classes]
corresponding course title, register to first receive course videos and materials, you can sign up for a single course or buy a card to participate in the whole year’s learning classes]
[Basic Section—For detailed introduction, please click on the image below corresponding course title, register to first receive course videos and materials, you can sign up for a single course or buy a card to participate in the whole year’s learning classes]
corresponding course title, register to first receive course videos and materials, you can sign up for a single course or buy a card to participate in the whole year’s learning classes]
[Clinical Research Section—For detailed introduction, please click on the image below corresponding course title, register to first receive course videos and materials, you can sign up for a single course or buy a card to participate in the whole year’s learning classes]
corresponding course title, register to first receive course videos and materials, you can sign up for a single course or buy a card to participate in the whole year’s learning classes]
To make it easier and more professional for everyone to do research in 2021, Medical Plus has launched three major types of VIP card services: SCI annual card, stored value card, and National Natural Science Foundation annual card:
1. Guaranteed SCI Annual Card
18,000/year, designated for one person, can participate in all Medical Plus courses except (National Natural Science Foundation application class, training camp), one expert one-on-one guidance on topics and articles
Team annual card 120,000/year, 10 people can use.
2. Stored Value Card
1. Pre-deposit 30,000, enjoy an 80% discount on all courses; deposit 50,000, enjoy a 75% discount on all courses
2. No limit on the number of people, times, or duration, until the funds are exhausted
3. National Natural Science Foundation Card
1. Youth
① 33,000/year, for one person, can participate in all Medical Plus courses (except training camp)
② Includes 15,000 youth application expert revision service
④ Includes five high-value application original documents
2. General
① 55,000/year, can allow two people to participate simultaneously in all(except training camp) courses
② Includes 20,000 general application expert revision service (limited to one person’s application revision)
④ Includes ten high-value application original documents

Good articles need your likes and shares~
↓↓↓↓