Click the blue text

Follow us
Introduction /Introduction
Fat mass and obesity-associated protein (FTO) is the first identified m6A demethylase, and its abnormal expression is associated with various cancers such as acute myeloid leukemia, melanoma, cervical cancer, and breast cancer. Developing highly sensitive and selective methods for FTO detection is crucial for early cancer diagnosis and drug development. In this regard, Professor Li Yan’s team at Northwestern University designed a DNAzyme-mediated CRISPR/Cas12a signal cascade amplification system and a carboxylated carbon nitride nanosheet/phosphorus-doped nitrogen vacancy modified carbon nitride nanosheet (C-CN/PCNV) heterojunction as the emitter for an electrochemiluminescence (ECL) biosensor, enabling highly sensitive and selective detection of the demethylase FTO.First, the C-CN/PCNV heterojunction was modified on the surface of a glassy carbon electrode (GCE) as an efficient ECL luminescent material, whose unique structure prevents electrode passivation and enhances electron-hole recombination efficiency. Subsequently, single-stranded DNA (ssDNA-Fc) labeled with ferrocene (Fc) was immobilized on the electrode surface to quench the ECL signal. When FTO is present, its demethylation activity activates the cutting activity of the DNAzyme, releasing primer P1 and triggering the trans-cleavage capability of the CRISPR/Cas12a system, which then cleaves ssDNA-Fc, restoring the ECL signal. This cascade amplification mechanism significantly improves the sensitivity and selectivity of detection, achieving ultra-low concentration detection of FTO.Related results were published under the title “Highly sensitive and selective demethylase FTO detection using a DNAzyme-mediated CRISPR/Cas12a signal cascade amplification electrochemiluminescence biosensor with C–CN/PCNV heterojunction as emitter” in Biosensors and Bioelectronics.
Research Figures /Introduction
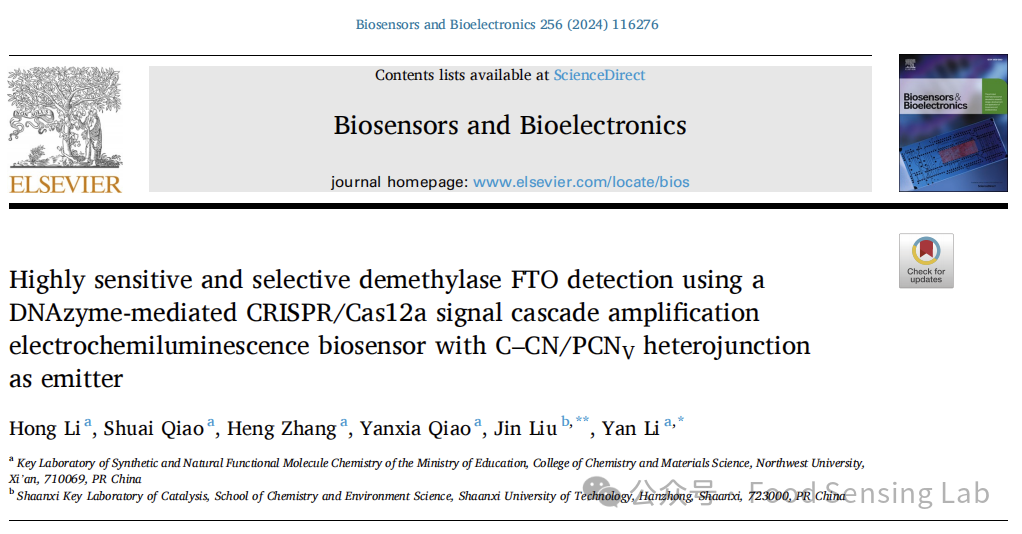
Scheme 1. (A) The synthesis of C–CN/PCNV heterojunction. (B) Fabrication of the ECL biosensor for FTO detection. (C) The m6A demethylation activated DNAzyme-mediated CRISPR/Cas12a process.

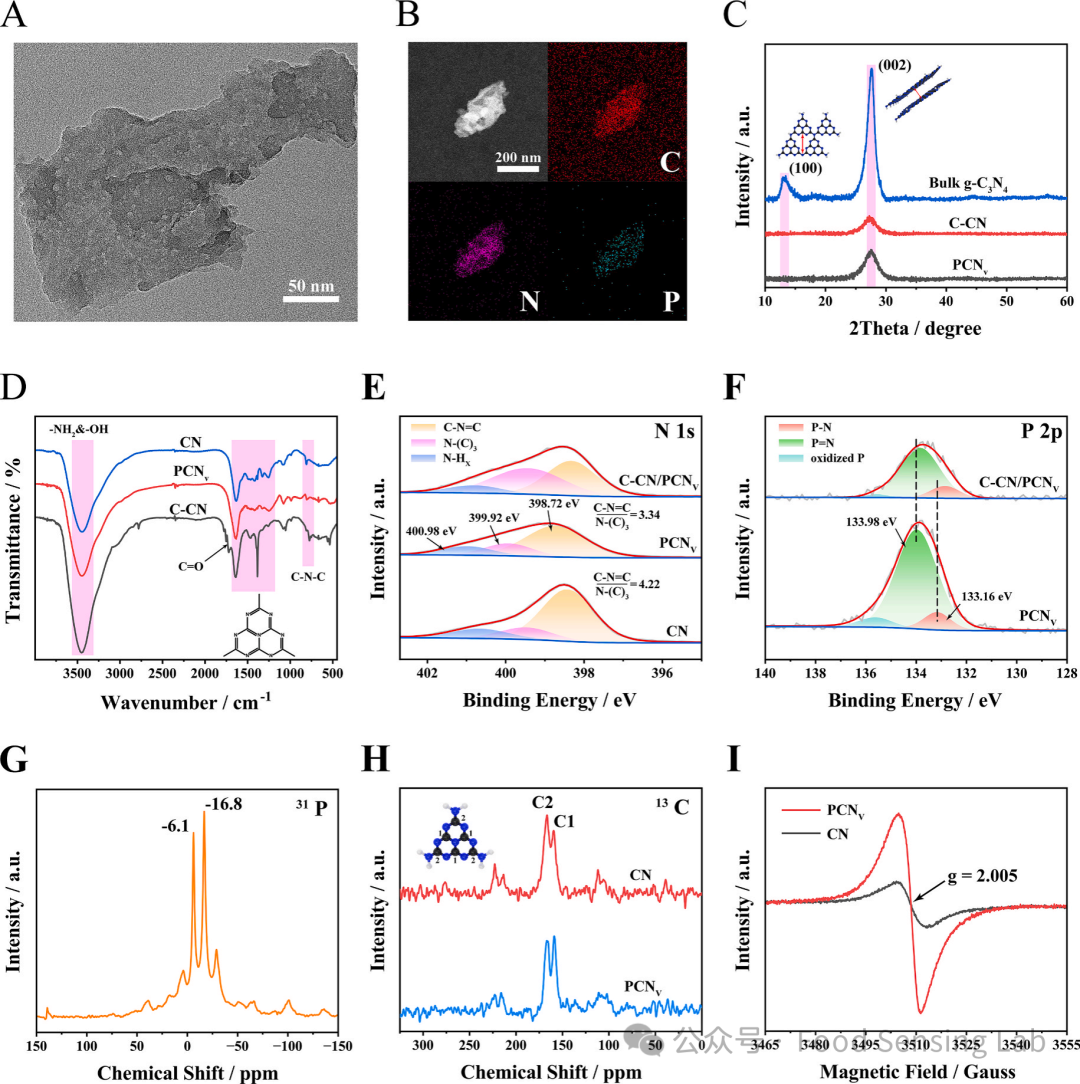
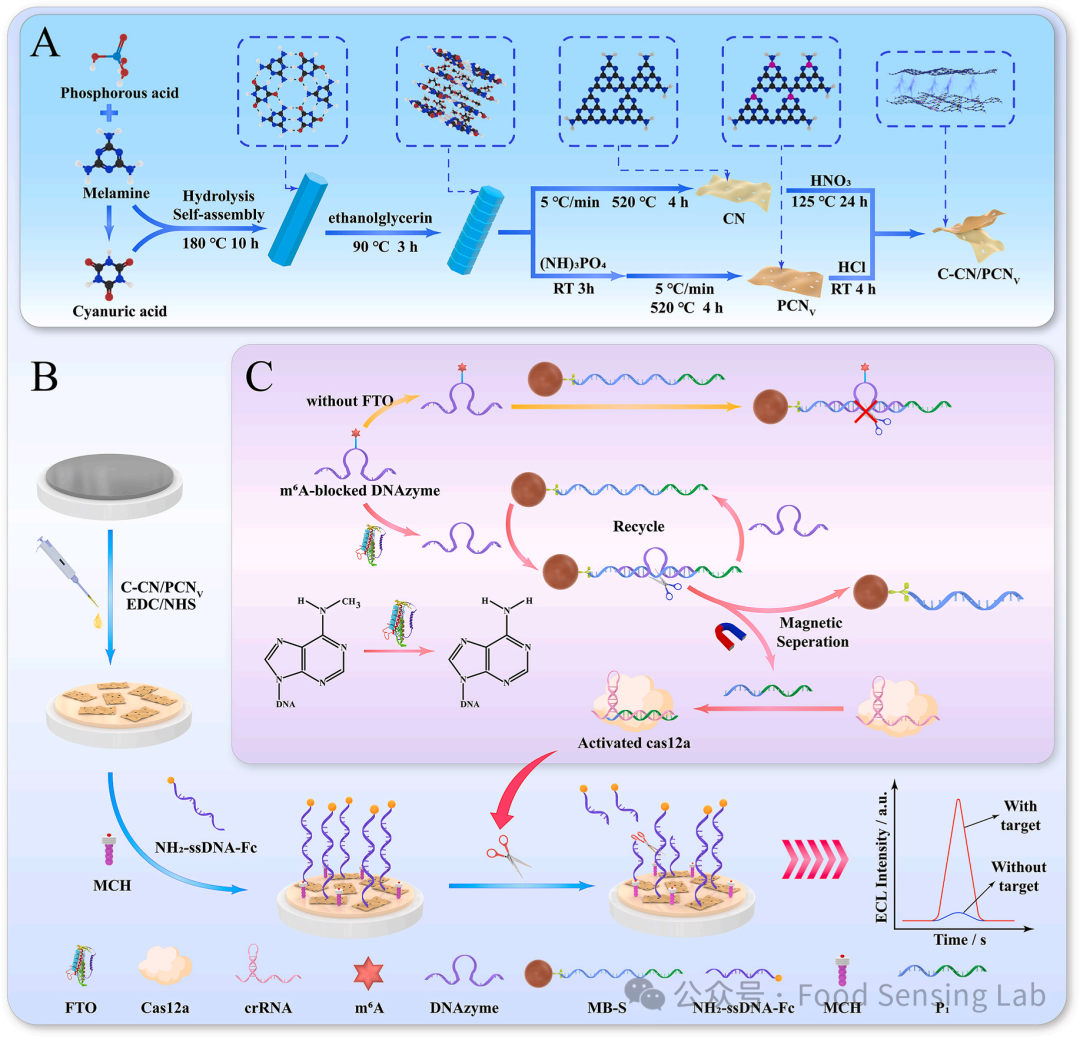
Fig 1. (A) TEM image of C–CN/PCNV heterojunction. (B) Element mapping image of C–CN/PCNV heterojunction. (C) XRD patterns of bulk g-C3N4, C–CN and PCNV. (D) FT-IR spectra of CN, C–CN and PCNV. High-resolution XPS spectra of (E) N 1s of CN, PCNV, C–CN/PCNV heterojunction and (F) P 2p of PCNV, C–CN/PCNV heterojunction. (G) 31P solid-state NMR spectra of PCNV. (H) 13C solid-state NMR spectra of CN and PCNV. (I) EPR spectra of CN and PCNV.

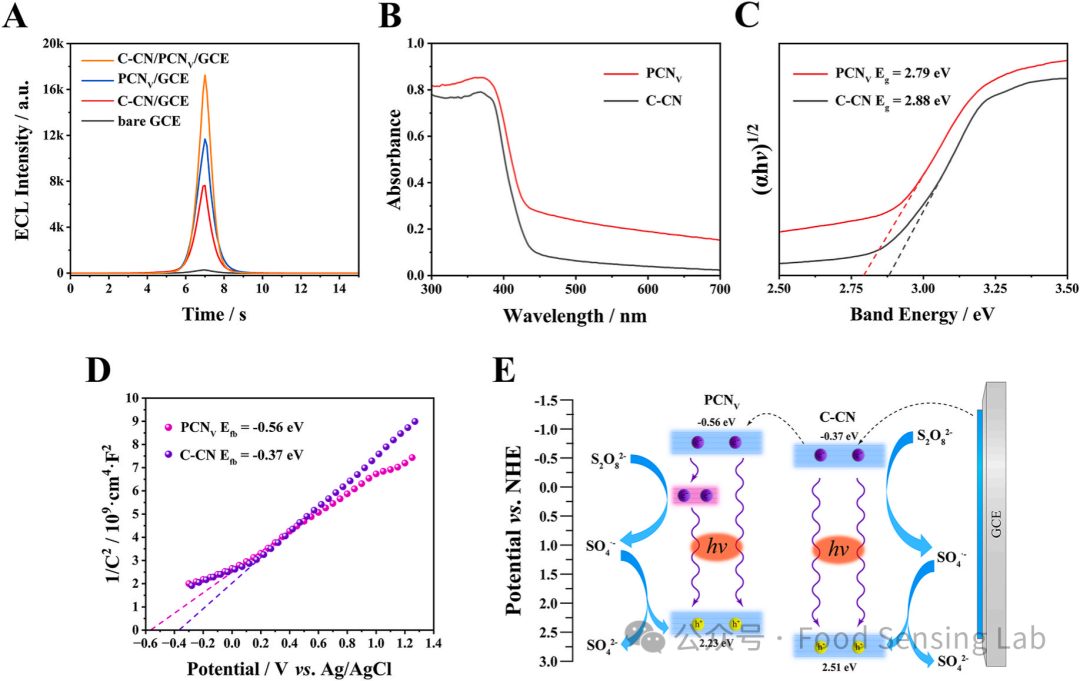
Fig 2. (A) ECL response of different electrodes. ECL signals were measured in 0.1 M PBS containing 50 mM K2S2O8 by scanning the potential from 0 to −1.4 V at a scanning rate of 100 mV s-1. The concentrations of C–CN, PCNV and C–CN/PCNV were 0.01 mg/mL, respectively. (B) UV–vis-DRS spectra of C–CN, PCNV. (C) Tauc plots of C–CN and PCNV. (D) Mott-Schottky plots of C–CN and PCNV. Measurement conditions: 0.1 M Na2SO4, frequency 3000 Hz. (E) ECL emission process of C–CN/PCNV heterojunction.

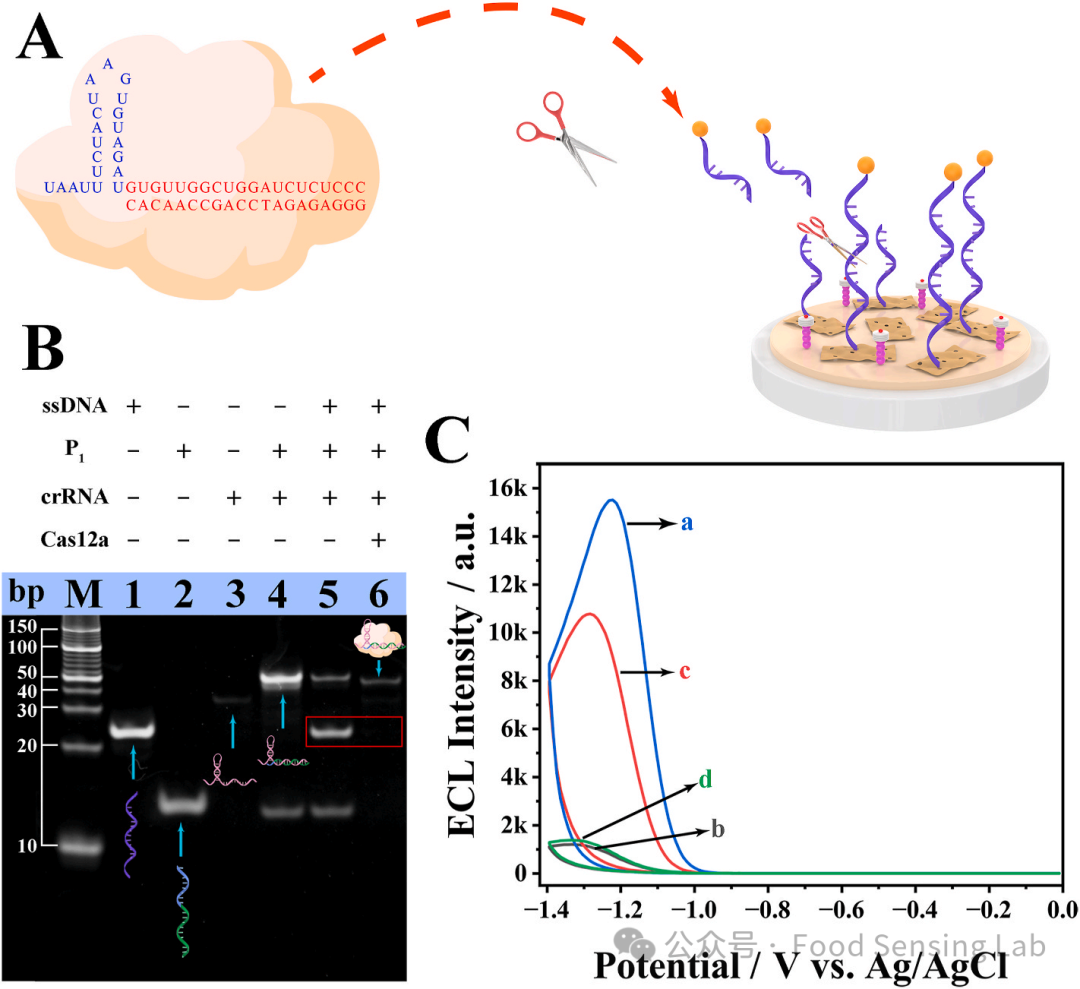
Fig 3. (A) Design of P1 and crRNA in the CRISPR/Cas12a system for the trans-cleavage of the ssDNA. (B) PAGE analysis of CRISPR/Cas12a cleavage reaction. M: DNA marker, Lane 1: ssDNA-Fc, Lane 2: P1, Lane 3: crRNA, Lane 4: crRNA + P1, Lane 5: ssDNA + P1 + crRNA, Lane 6: ssDNA + P1 + crRNA + Cas12a. (C) ECL intensity vs potential profiles of different electrodes. (a) C–CN/PCNV/GCE, (b) ssDNA-Fc/C–CN/PCNV/GCE, (c) ssDNA-Fc/C–CN/PCNV/GCE interaction with activated CRISPR/Cas12a, (d) ssDNA-Fc/C–CN/PCNV/GCE interaction with inactivated CRISPR/Cas12a (a scrambled P1 sequence that cannot bind to crRNA). ECL signals were measured in 0.1 M PBS containing 50 mM K2S2O8 by scanning the potential from 0 to −1.4 V at a scanning rate of 100 mV s-1 and PMT = −600 V. The concentrations of C–CN/PCNV: 0.01 mg/mL, ssDNA-Fc:1 μM, FTO: 100 nM.

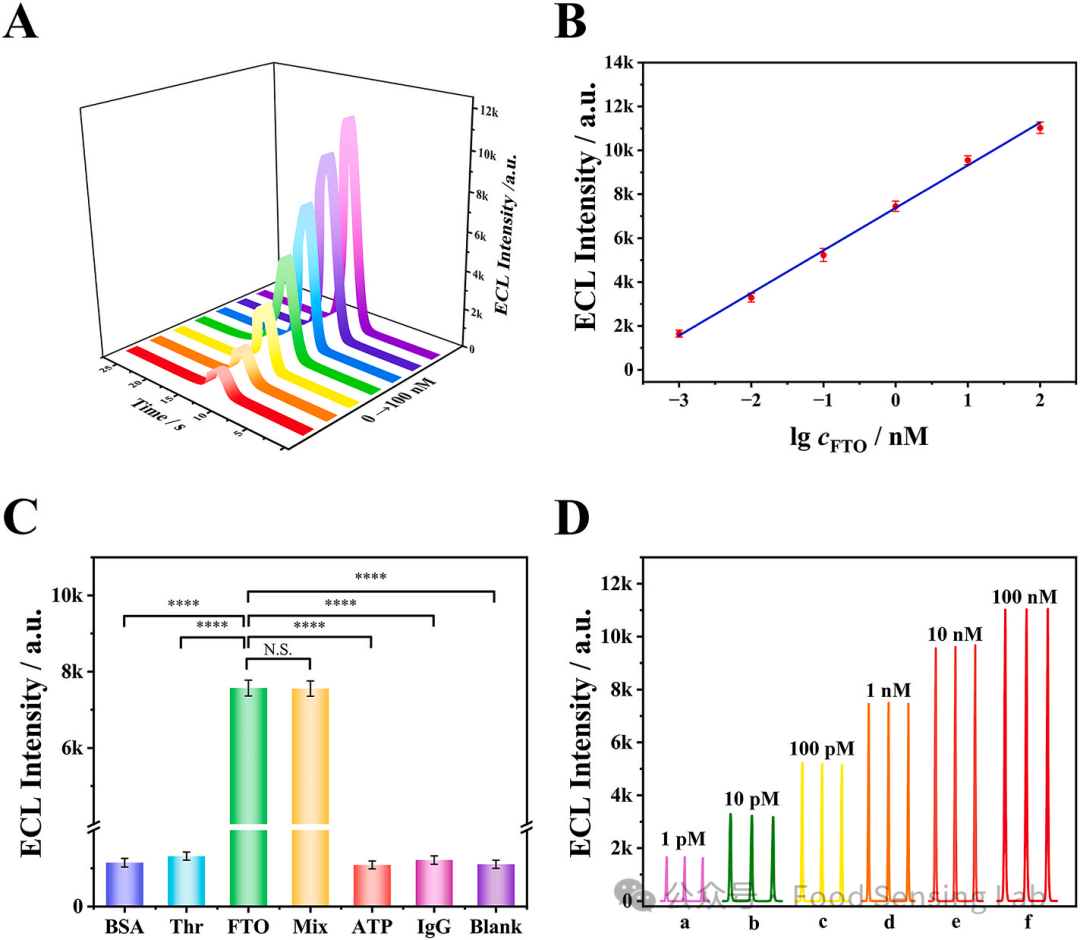
Fig 4. (A) ECL intensity−time curves of the biosensor incubated with different concentrations FTO: 0, 1 pM, 10 pM, 100 pM, 1 nM, 10 nM, 100 nM. (B) Corresponding relationship between the ECL intensity versus logarithm of FTO concentrations. (C) ECL response of the biosensor for the detection of 1 nM FTO, 50 μg/mL BSA, 100 nM Th, 5 nM ATP and 100 nM IgG. Error bar represents SD (n = 3). ****P value < 0.0001. N.S., no significance. (D) Repeatability with different concentrations of FTO from 1 pM to 100 nM. Measurement conditions are the same as Fig. 3.

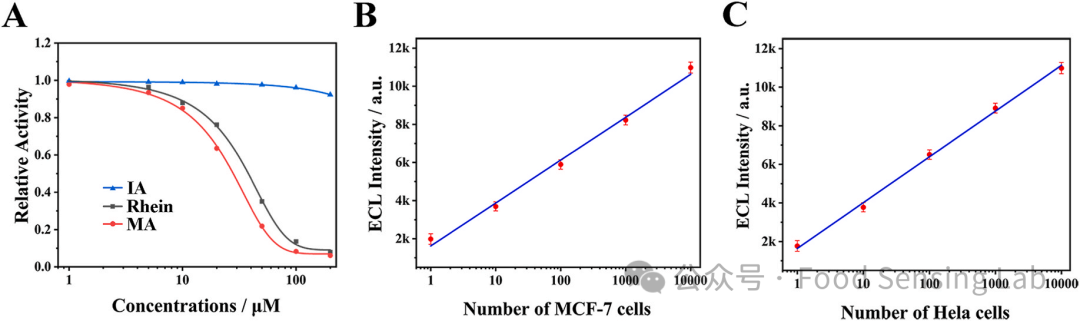
Fig 5. (A) Relative activities of FTO induced by different concentrations of rhein (green curve), MA (red curve), and negative control IA (blue curve). The concentration of FTO was 100 nM. Linear relationship of ECL response with the logarithm of MCF-7 cell (B) and Hela cell (C) that the cell number from 1 to 10,000 cells. Measurement conditions are the same as Fig. 3.

Conclusion /Conclusion
In summary, this study constructed a heterojunction using carboxylated carbon nitride nanosheets (C-CN) and phosphorus-doped nitrogen vacancy modified carbon nitride nanosheets (PCNV) as the ECL emitter, significantly enhancing electron-hole recombination efficiency and preventing electrode passivation. Through the DNAzyme-mediated CRISPR/Cas12a signal cascade amplification system, the presence of FTO activates the cutting activity of the DNAzyme, generating activated DNA, which in turn activates the trans-cleavage capability of Cas12a, cleaving the ssDNA-Fc probe and restoring the ECL signal. This biosensor exhibits high sensitivity for FTO detection in the range of 1.0 pM to 100 nM, with a detection limit as low as 0.63 pM, and has been successfully applied to the detection of FTO in cancer cell lysates and inhibitor screening, demonstrating great potential for early clinical diagnosis and drug discovery, providing a new approach for the detection of cancer-related biomarkers.
Literature Information /Literature information
LI H, QIAO S, ZHANG H, et al. Highly sensitive and selective demethylase FTO detection using a DNAzyme-mediated CRISPR/Cas12a signal cascade amplification electrochemiluminescence biosensor with C–CN/PCNV heterojunction as emitter [J]. Biosens Bioelectron, 2024, 256: 116276.
https://doi.org/10.1016/j.bios.2024.116276

Scan the QR code above
Follow us

END

For submissions, recommendations, or collaborations, please contact the email