PRGdb: Overview of the Plant R Gene Database
Background Introduction
Nearly 40% of global crop yields are lost due to pests and diseases, and plant breeders and researchers have made significant efforts to identify genes related to plant disease resistance mechanisms. Plants have evolved the ability to recognize potential pathogens and predators, activating defense mechanisms to counter them. The activation of these mechanisms is based on specific receptors encoded by Pathogen Recognition Genes (PRGs). However, knowledge of plant-pathogen interactions is often derived from studies of individual genes, while disease responses are co-regulated by highly interconnected gene networks and various processes and pathways.
Despite the diversity in PRGs, the leucine-rich repeat (LRR) domain is ubiquitous. This domain is found in Pattern Recognition Receptors (PRRs) as transmembrane proteins, which recognize external signals and trigger the first layer of induced defense (also known as pathogen-associated molecular pattern-triggered immunity or PTI). These can be divided into two main categories: RLP, which contains only LRR and transmembrane (TM) domains; and RLKs, which contain a kinase (KIN) domain. Additionally, LRR domains are present in NLR proteins, which also contain a nucleotide-binding site (NBS) domain. These receptors trigger a stronger immune response (effector-triggered immunity or ETI) and can be further classified into two main types: TNL, which also contains a Toll-Interleukin 1 receptor (TIR) domain; and CNL, which carries an additional coiled-coil (CC) domain. There are also receptors with other domains besides the LRR domain.
So far, extensive transcriptomic studies have been conducted in the field of plant-pathogen interactions, establishing transcriptomics as a suitable platform to elucidate the complexity of the molecular mechanisms underlying these interactions. Despite progress in the fields of omics and bioinformatics, exploratory data analysis remains a tedious task, and studying PRGs using bioinformatics tools poses challenges for a significant portion of the scientific community. The PRGdb database was developed to fill this gap, providing references for genes involved in plant disease resistance processes.

Brief Introduction to PRGdb
The new version of the Plant Resistance Gene Database (PRGdb; http://prgdb.org/prgdb4/) has been expanded to keep pace with the growing amount of available knowledge and data (proteomics sequencing, cloned genes, publicly analyzed data, etc.). The database website has updated prediction tools, increased data, and new sections. The new sections include plant resistance transcriptomic experiments, providing additional easily accessible experimental information. As an automated annotation and prediction tool for plant resistance genes behind PRGdb, DRAGO3 has improved accuracy and sensitivity, making the prediction results more reliable. PRGdb has obtained 199 reference resistance genes and 586,652 inferred resistance genes from 182 sequenced proteomes.
Compared to previous versions, PRGdb 4.0 has increased the number of reference resistance genes from 153 to 199 and the number of inferred resistance genes from 177K in 176 proteomes to 586K in 182 proteomes. It integrates public transcriptomic data from studies focusing on plant-pathogen interactions in five agricultural species. In version 4.0 of PRGdb, the types of PRGs have expanded to seven types, including LYK, LYP, and LECRK receptors. The annotation tools, disease resistance analysis, and gene orthology (DRAGO) have also been improved to provide more accurate and sensitive annotations for any given DNA or amino acid (AA) sequence.
The creation of PRGdb 4.0 can provide references for the global plant science community and breeders, aiding further research into plant resistance mechanisms against pathogens. Interested parties can browse at http://prgdb.org/prgdb4/.

Main Improvements in the New Version
Established New Reference PRG Types and Proteomes
Retrieved new cloned resistant genes, expanding search targets beyond LRR receptors to include cloned genes for LYK, LYP, and LECRK proteins. The structural domain composition was confirmed using public prediction tools such as InterProScan, Pfam, CDD, Smart, and Prosite, establishing new reference PRGs. A total of 51 proteins were included as reference PRGs in the new version.
Established HMM
Reference gene AA sequences from the seven classes of resistant genes (CNL, TNL, RLK, RLP, LYK, LYP, and LECRK) included in PRGdb 4.0 were isolated. Multi-sequence alignments (MSA) were constructed for each category using MEGA X. Hidden Markov Models (HMMs) were built using the MSA. HMMs for each domain were constructed from the MSAs of each category, except for the LYSM domain (where LYK and LYP proteins were combined to generate additional HMMs for LYS and LECM) and LECM (two subgroups within LECRK also constructed additional HMMs: one containing the legume LECM domain and another containing the globular LECM domain).
Using InterProScan (activating SMART, Pfam, CDD, and Prosite tools) to locate resistance domains within the MSA, and using MEGA software to visualize the MSA and determine the origins of the HMMs. HMMs were tested against the original FASTA files using hmmsearch (hmmer tools; http://hmmer.org/) to verify their utility in predicting resistance domains.
New DRAGO3 Features
Alignment scores for different hits were calculated based on the BLoSuM62 matrix. HMMs were updated (except for CNL and TNL classes), and three new protein classes were added: LYK, LYP, and LECRK, along with all other non-standard domain combinations. A minimum score threshold for DRAGO3 was defined, and the same predictions were conducted for CC and TM domains using COILS 2.2 and TMHMM 2.0 software, respectively.
The Arabidopsis proteome annotation from Araport 11 was analyzed using DRAGO2 and DRAGO3, and the inferred resistance genes were analyzed using InterProScan, comparing the results with two criteria: proteins predicted by both DRAGO and InterProScan; proteins predicted by DRAGO that are the same or better.
The most conserved regions from the MSA of each resistance level were extracted to construct 209 HMMs. These HMMs were further filtered based on the following criteria: HMMs belonging to unrelated regions (i.e., TM domains, which were analyzed using the TMHMM tool rather than DRAGO3 HMMs); HMMs that could not identify the proteins used to construct them; new HMMs were compared with DRAGO2 HMMs, retaining the higher-performing HMMs.
A total of 109 HMMs were retained, nearly double that of the previous DRAGO2.
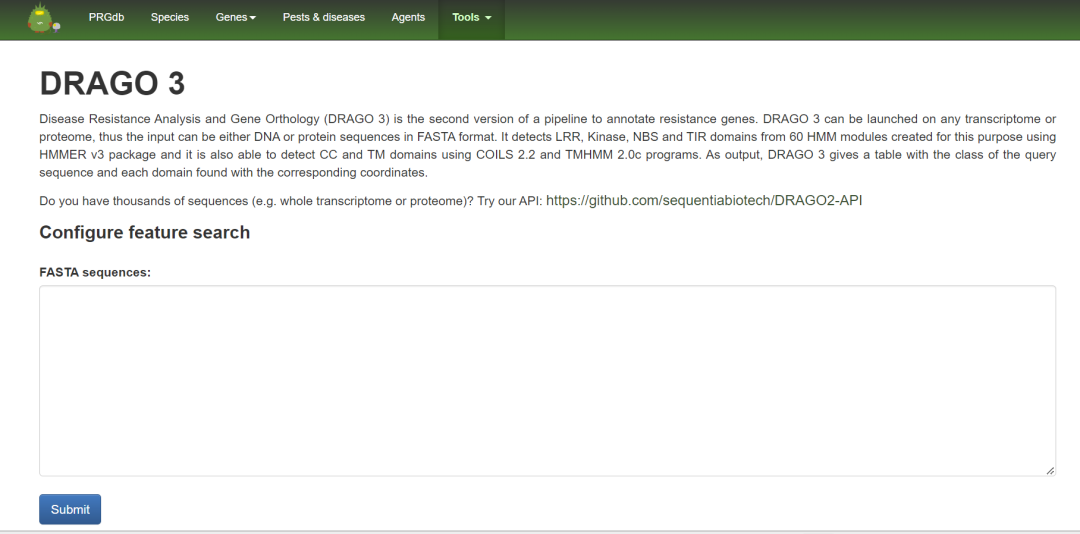
Public RNA-seq Experimental Data
Publicly available RNA-seq experiments were conducted to study the interactions of plants with five agricultural crops: tomato, rice, wheat, grape, and Arabidopsis. In these studies, different plant species were challenged by various pathogens, such as bacteria, fungi, insects, and viruses. The list of DEGs for rice, wheat, grape, and Arabidopsis was retrieved from the literature. For tomato, the original sequencing data (fastq files) were downloaded from NCBI’s SRA repository (https://www.ncbi.nlm.nih.gov/sra) using the SRA-toolkit (http://ncbi.github.io/sra-tools/), and analyzed on the same bioinformatics pipeline using the web-based A.I.R. RNAseq analysis package (https://transcriptomics.sequentiabiotech.com/) to explore and compare different studies.
Additionally, the new database underwent new annotations, incorporating three new resistance categories: LYK, LECRK, and LYP, bringing PRGdb 4.0 to include seven typical resistance protein categories in plants, and it can predict domain combinations beyond these established categories. PRGdb 4.0 includes a total of 199 reference resistance genes. By utilizing DRAGO3 to analyze 182 plant proteomes and their inferred resistance genes included in prgdb4.0, RLK and RLP remain the most abundant categories, while resistance classes LECRK, LYP, and LYK are the least abundant.
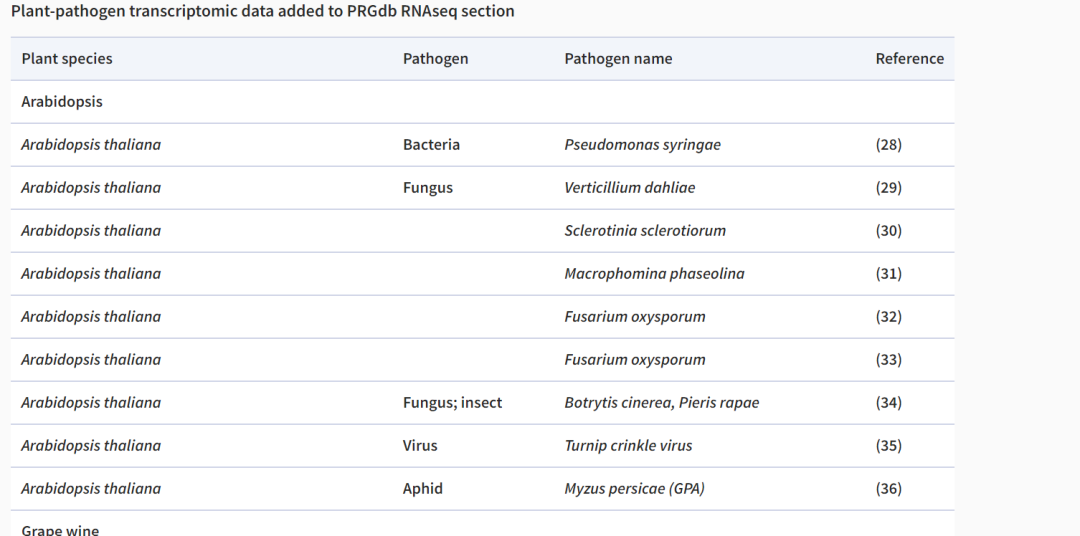
Through plant-pathogen transcriptomic analysis, 35 RNA-seq studies were obtained, resulting in a list of differentially expressed genes (DEGs) for all these genes, which were incorporated into the new section of PRGdb4.0.
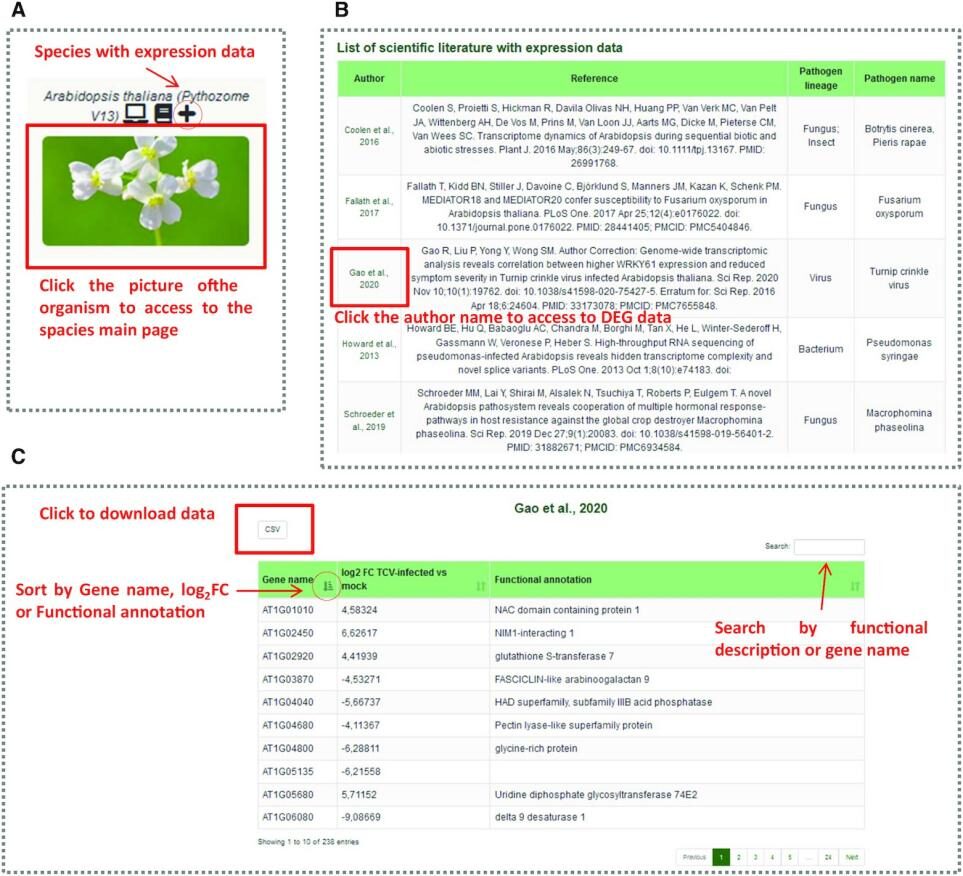
This section is easily accessible via pages providing expression analysis for the five species (Figure A). Available experiments for specific species are displayed on the homepage (Figure B). Selecting one will take users to the DEG matrix (Figure C). Users will be able to explore the results of differential expression analysis from various studies. This information is displayed as a DEG matrix of upregulated and downregulated genes, reporting gene IDs, log2 fold changes, and gene functional annotations for comparison of interest and gene functions across studies. Users can download the data in csv format, which can be sorted by gene ID, log FC, and functional description. Experimental perspectives can be provided for genes of interest.
The new version of PRGdb 4.0 enhances utility in the fields of plant science and breeding. It can be used to query resistance genes from many plants and algae, analyze the sequences of resistance genes to predict resistance genes, and study gene expression under specific plant pathogen conditions. The addition of new proteomes makes previously impossible species studies feasible. With new reference PRGs and genomic data being made public, PRGdb will continue to integrate multi-omics data, becoming a reference database for plant researchers to address key challenges in plant-pathogen interactions.
Interested Readers Can Refer to the Following Literature:
Calle García, J., Guadagno, A., Paytuvi-Gallart, A., Saera-Vila, A., Amoroso, C. G., D’Esposito, D., Andolfo, G., Aiese Cigliano, R., Sanseverino, W., & Ercolano, M. R. (2022). PRGdb 4.0: an updated database dedicated to genes involved in plant disease resistance process. Nucleic acids research, 50(D1), D1483–D1490.
Tan, Y. C., Kumar, A. U., Wong, Y. P., & Ling, A. P. K. (2022). Bioinformatics approaches and applications in plant biotechnology. Journal, genetic engineering & biotechnology, 20(1), 106.
End of Article Promotion
Highly recommend you share with nearby postdoctoral researchers and young biology PIs to enhance their research with more data insights:
-
2024 Shared Server Friend Price is still 800 -
Finally Available Exclusive Bioinformatics Cloud Server