Today, let’s talk about the Plant Resistance Gene Database (PRGdb).
What is an R gene? It is a plant’s disease resistance gene (Plant Resistance Gene). Unlike animals that have immune cells in their immune system, plants do not have their own immune cells. So how do plants combat pathogen invasion? First, let’s take a look at the immune mechanisms that plants use to respond to pathogens:
In response to pathogen invasion, plants primarily have two stages of defense mechanisms: Broad-spectrum defense PTI (PAMP-triggered immunity) and Specific defense ETI (effector-triggered immunity).
The specific process is as follows: when pathogens infect plants, certain pathogen-associated molecules such as bacterial lipopolysaccharides, flagellin proteins, lipopeptides, and peptidoglycans, collectively known as pathogen/microbe-associated molecular patterns (pathogen/microbe associated molecular pattern, PAMP/MAMP), are recognized by corresponding pattern recognition receptors (pattern recognition receptors, PRR) in plants, triggering a broad-spectrum defense response that inhibits the initial infection by the pathogen. Then, the pathogen counters by using specific effector proteins (effector) to suppress the plant’s PTI, aiding the pathogen’s reproduction or spread within the plant. At this point, plants cannot back down; in response to the specific effector proteins of the pathogen, some plants have evolved resistance genes (R genes) to recognize these effector proteins, thereby triggering the plant’s hypersensitive response, leading to cell apoptosis and preventing further spread of the pathogen. This response is known as effector-triggered immunity (effector triggered immunity, ETI) (Figure 1).
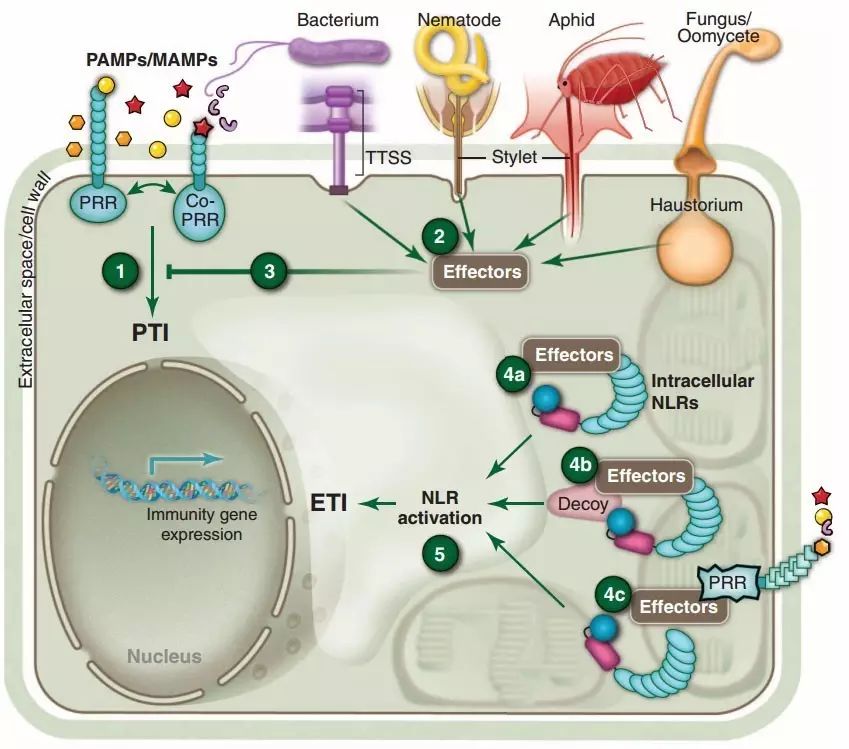
Figure 1 Mechanism of plant resistance to pathogen invasion
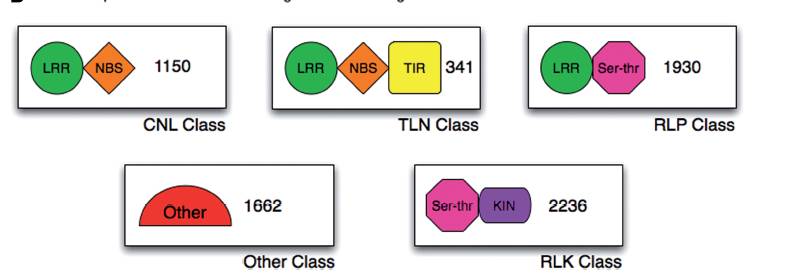
The PTI and ETI in plants are analogous to innate and adaptive immunity in animal cells, with one being broad-spectrum and the other being specific. Among them, plant R genes play a crucial role in ETI. R genes generally have several specific domains, and different types of R genes can have different combinations of these domains. For example, the common structure of R genes includes a leucine-rich repeat region (leucine-rich repeat, LRR), a nucleotide-binding domain (nucleotide-binding, NB), and TIR (Toll/interleukin-1-receptor).
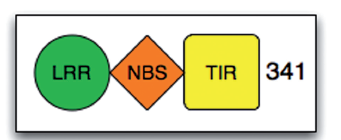
These different combinations of domains form various types of R genes, as shown in the following figure:
The Plant Resistance Gene Database (Plant Resistance Gene database, PRGdb), http://prgdb.crg.eu/wiki/Main_Page, is a public database concerning plant R genes. The latest version of this database (from 2013) includes 106,373 R genes from 241 plant species and will continue to be updated and enriched. These R genes have five sources:
-
Collected from existing literature;
-
Collected from NCBI GenBank;
-
Predicted from NCBI UniGene;
-
Predicted from the Pythozome database;
-
Uploaded by other researchers.
PRGdb contains 10 types of domains, including MLO, PRW8, GNK2, CC, TIR, NBS, LRR, Receptor-Like Kinase, Ser-Thr Kinase, and others. Additionally, PRGdb also includes 138 types of pathogens with 23 effector genes (avirulence genes) and 120 disease information entries.
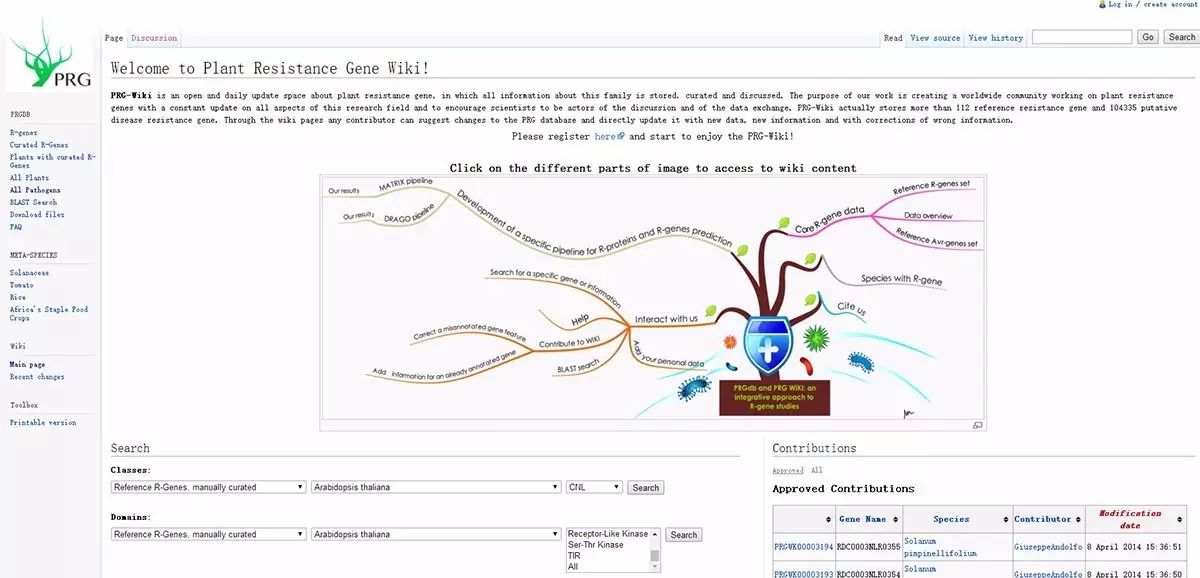
Next, let’s see how to search for R genes of interest using PRGdb:
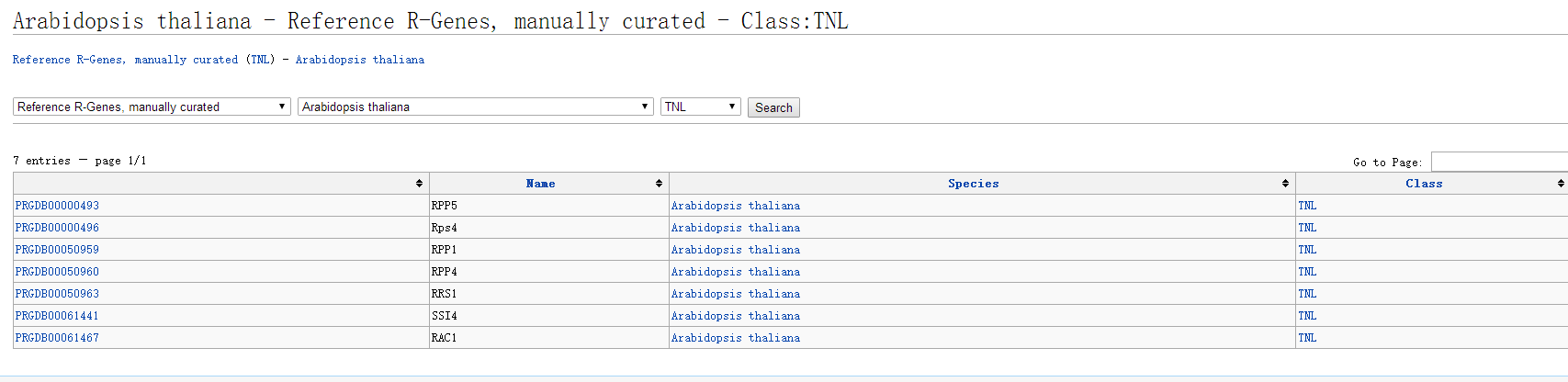
PRGdb provides two methods to search for R genes: searching by R gene type and searching by the domains contained in R genes. For example, if we want to know what TNL type R genes have been reported in Arabidopsis, we can set the search criteria from the “Search” section at the bottom of the database homepage, selecting “Reference R-Gene, manually curated”, “Arabidopsis thaliana”, and “TNL”, then click “search”.

Seven genes were found:
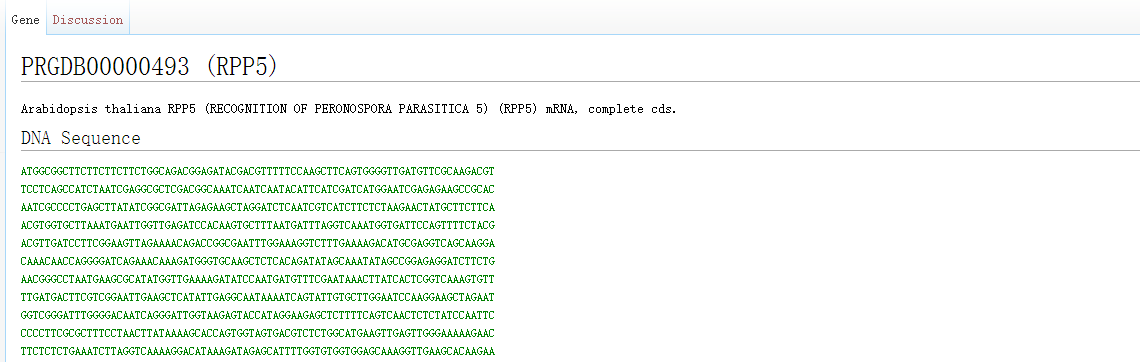
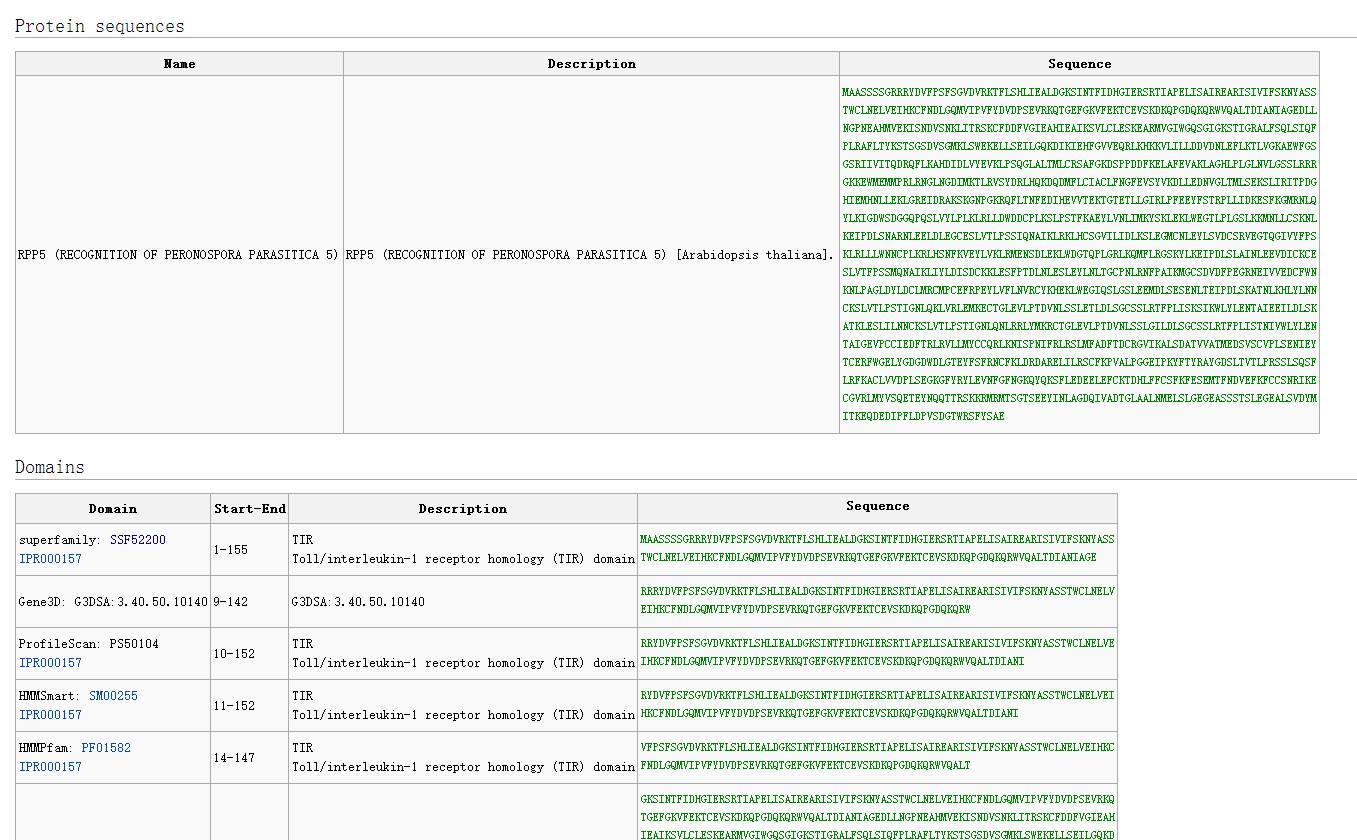
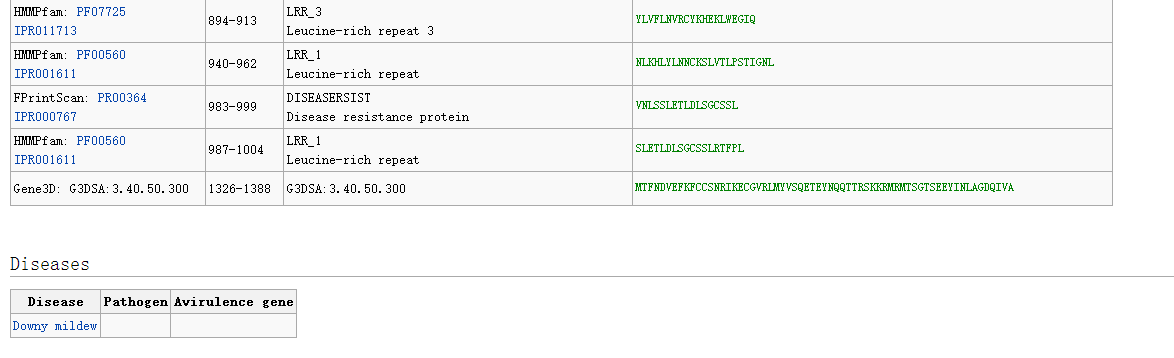
By clicking on the gene name, we can obtain detailed information about the R gene, including CDS sequence, amino acid sequence, gene function description, domain location and sequence, diseases, etc.
If you have a bunch of unigene and want to know if there are any R genes among them, you can also perform a BLAST comparison in PRGdb to search for R genes.
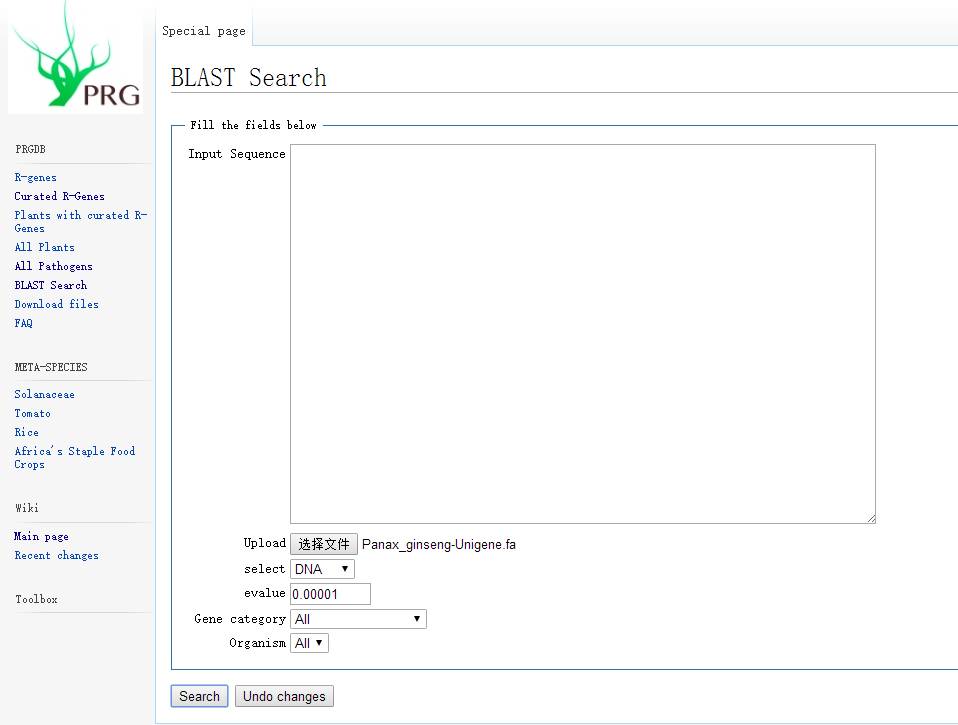
However, I tested it and I don’t know what the file size limit is; I tried a 37MB fasta file and it failed; uploading half the data also failed, which was quite frustrating…
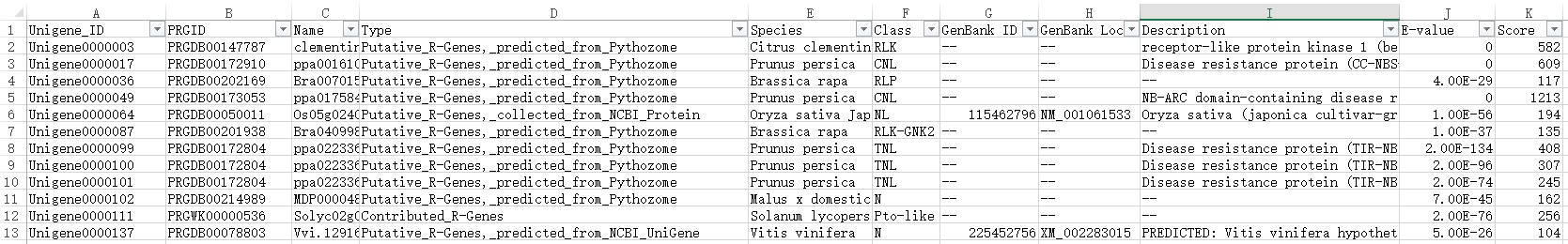
Is there a method to predict R genes from a large number of unigene? Of course! The transcriptome sequencing provided by Gidiao offers annotations for the plant disease resistance gene database, providing detailed R gene annotation results, including R gene names, types, species, annotations, etc., along with classification histograms that visually display the number of various types of R genes, saving time and effort!
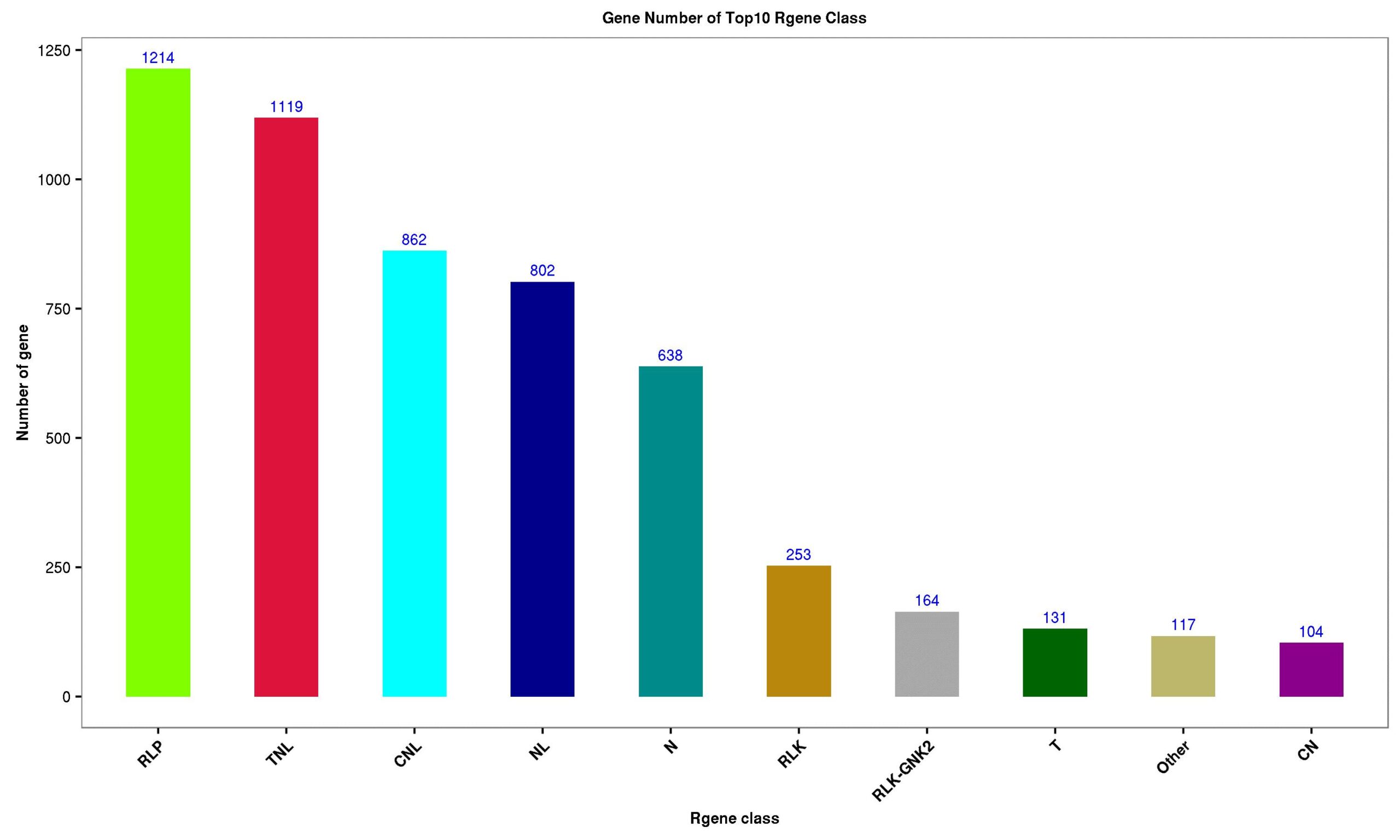
Finally, I want to emphasize that research on plant R genes plays a significant role in elucidating the molecular mechanisms of plant resistance to pathogens and in the breeding improvement of crops. For example, we can transfer the disease resistance gene from Arabidopsis into tomatoes, enabling tomatoes to acquire resistance to certain diseases, thus producing disease-resistant tomatoes. If you want to learn more about plant disease resistance and R genes, you can participate in our online class next Wednesday on “Applications of Transcriptome Sequencing in Pathogen-Host Interactions“.
Class content 
-
Analysis of the principles of pathogen-host interactions
□ Molecular mechanisms of pathogen-host interactions
□ Interpretation of related pathways in KEGG
□ Analysis of R gene types and structures
-
Key points in project design for Dual RNA-seq applications in this field
□ How to choose library construction strategies
□ What is a reasonable sequencing amount
□ What issues should we pay attention to in sampling
□ What customized bioinformatics analysis strategies can help conduct such research
Communication time:
December 21 (next Wednesday) at 16:00
Guest speaker:
Teacher ZhouRegistration link:
https://ke.qq.com/course/141293#tuin=8e8efa7

Literature interpretation/technical posts/tutorials/frontier information,
Sharing a piece of valuable content with you every day~
We aim to be the most informative public account in the industry!
Our dream is in the future,
Gidiao can help you reach higher levels in the field of scientific research!