The endonucleases with sequence-specific catalytic characteristics have become important tools for nucleic acid target identification, especially the CRISPR/Cas system. However, the diagnostic applications of the CRISPR/Cas system still face several challenges, including increased costs and instability associated with the use of guide RNA. To address these limitations, researchers are actively exploring and developing alternative strategies.
Argonaute (Ago) proteins play a key role in RNA interference (RNAi) and small RNA-guided gene regulation pathways. They can cut target nucleic acids without needing specific sequence motifs, and Ago can precisely cleave complementary targets between nucleotides 10 and 11 of the guide DNA, allowing for less restrictive nucleic acid selection. In current research, Ago proteins are primarily used for direct cleavage after nucleic acid amplification reactions, such as PCR, loop-mediated isothermal amplification (LAMP), etc., producing enhanced fluorescent signals. However, these methods require more enzymes and procedural steps, leading to relatively complex processes. Meanwhile, some research teams have designed methods using Ago proteins without nucleic acid amplification, but the performance is relatively poor.
Recently, a research team published an article titled “ANCA: artificial nucleic acid circuit with argonaute protein for one-step isothermal detection of antibiotic-resistant bacteria” in the journal Nature Communications. In this article, the authors proposed a DNA detection technology that combines an artificial nucleic acid circuit with Ago protein cleavage activity (ANCA), establishing a high specificity and exponential signal amplification positive feedback loop. This reaction is a one-step process and can easily detect different targets by changing the nucleic acid recognition sites. The authors used the ANCA method combined with a 3D nanocolumn swab to directly identify the ability to recognize bacteria producing carbapenemase from surfaces such as pig skin, tables, gloves, scissors, handles, and tweezers. The ANCA detection was applied to rectal swab specimens for diagnosing patients infected with carbapenemase-producing Klebsiella pneumoniae (CPKP), achieving clinical sensitivity and specificity of 100% and 100%, respectively.

Image source: Nature Communications

Main Content
Design of ANCA Method
The ANCA experiment utilizes two characteristics of Ago proteins. First, the cutting sites of Ago proteins are located at the 10th and 11th positions of the 5′ end of the guide DNA. Second, the DNA strands cleaved by Ago proteins can serve as guide DNA. The authors established a cycle using these two characteristics to develop the ANCA method (as shown below). The authors designed guide DNA 1 (G1), guide DNA 2 (G2), a reporter gene (R), and a complementary sequence of the reporter gene (R*). All sequences were designed to ensure that Trigger 1 (T1) and Trigger 2 (T2), generated during the reaction, can also serve as guide DNA.
The Ago/G1 and Ago/G2 complexes hybridize with complementary target DNA sequences, cleaving the respective targets, producing short DNA fragments (T1) from the target DNA. T1 can serve as another guide DNA, forming the Ago/T1 complex, which recognizes and cleaves R, resulting in the release of two short DNA strands (Output and T2). The output generates a fluorescent signal, and T2 again recognizes and cleaves R*, producing T1 and completing the DNA cycle. Through this positive feedback loop, repeated cleavage reactions can occur in the presence of target DNA, ultimately determining the presence of target DNA in the sample by monitoring the fluorescent signal.
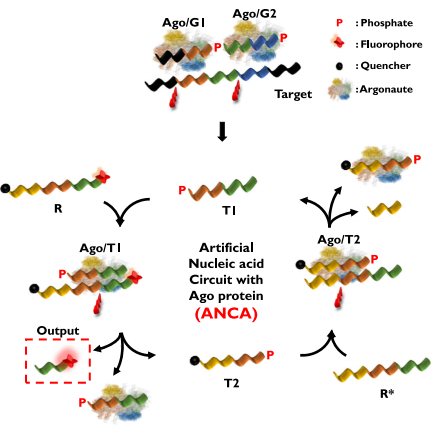
Schematic diagram of ANCA method detecting target nucleic acids. Image source: Nature Communications
Evaluation of ANCA Method
The ANCA method was designed to provide exponential amplification through a positive feedback loop (Figure a). The authors first evaluated different components of the ANCA method targeting Klebsiella pneumoniae carbapenemase (KPC). Strong fluorescent signals were observed during the ANCA reaction process in the presence of all reaction components (target DNA, G1, G2, Ago protein), and bands of the degraded R-R* structure were observed (Figure c). In the absence of any reaction component, the fluorescent signal was negligible, and the intact R-R* bands were observed.
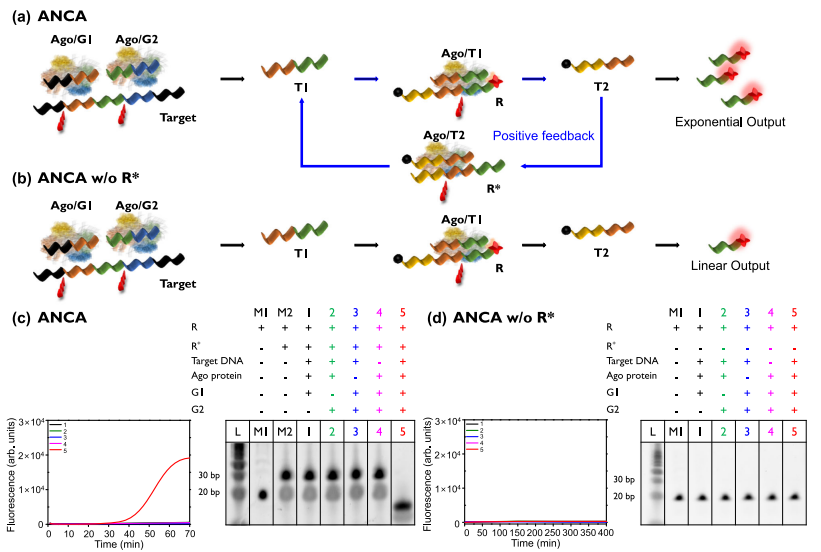
Evaluation of the ANCA method. Image source: Nature Communications
Optimization and Sensitivity of ANCA Method
The authors optimized the reaction temperature and concentrations of R, R*, Ago protein, and Mg2+. The optimal detection conditions were a reaction temperature of 75℃, concentrations of R and R* at 500 nM, Ago protein concentration at 200 nM, and MgCl2 concentration at 10 mM. Additionally, the concentrations of NaCl and bovine serum albumin (BSA) in the enzyme storage buffer were optimized to 75 mM and 10µg/mL, respectively, and these conditions were used for subsequent experiments.
Under the optimized conditions, the sensitivity of the ANCA method was evaluated using KPC and imipenemase (IMP) as target sequences. Figure a shows the time-dependent fluorescent intensity of different concentrations of KPC sequences. The threshold time was determined as the reaction time at which the fluorescent intensity reached 10,000. A linear relationship was observed in the range of 10 fM to 10 nM, with a limit of detection (LOD) of 1.87 fM. The ANCA method was used to detect IMP, revealing a linear relationship from 1 fM to 1 nM, with an LOD calculated as 178 aM (Figures c, d). Similarly, the authors designed nucleic acid circuits for detecting other carbapenemases such as VIM, NDM, and OXA-48 and evaluated their sensitivity. The results showed that the ANCA method has flexibility and broad target applicability, successfully detecting KPC, IMP, VIM, NDM, and OXA-48 sequences with slight modifications to the ANCA method.
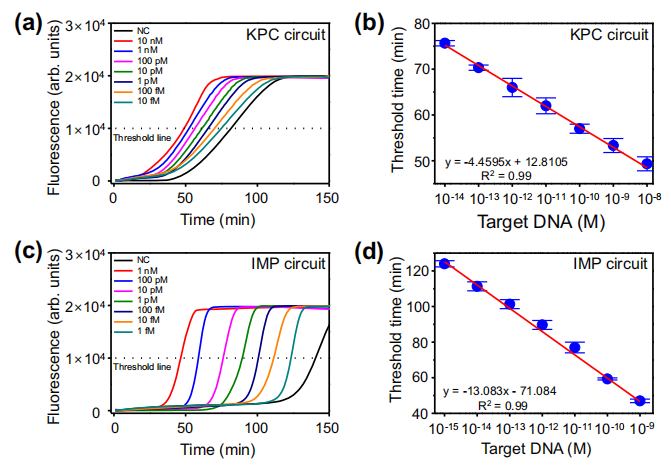
Sensitivity of the ANCA method. Image source: Nature Communications
Direct Detection of Resistant Bacteria Using ANCA Method
The authors attempted to detect carbapenemase-producing Klebsiella pneumoniae (CPKP) using the ANCA method without DNA purification. The resistant bacteria were directly added to a tube containing the ANCA reaction mixture and incubated at 75℃. The results confirmed that the ANCA reaction directly detected the target antibiotic-resistant bacteria (Figures b and c) using KPC and IMP. The authors also attempted to use VIM, NDM, and OXA-48 circuits to detect CPKP, finding that each circuit could effectively distinguish WT and CPKP. The ANCA method allows for simplified detection of CPKP without pre-concentration, lysis, and DNA purification steps, attributed to the fact that bacteria can be lysed within 15 minutes at temperatures above 60℃.
To explore the application of the ANCA method in clinical specimen bacterial detection, the authors added bacteria to urine and blood samples, directly mixing them with the ANCA reaction mixture and incubating. The KPC circuit was able to detect KPC in 99% of human urine (Figure e). The IMP circuit also successfully detected Klebsiella pneumoniae (IMP) in urine (Figure f). Additionally, the ANCA method identified KPC and IMP in 90% and 99% of blood samples, respectively (Figures h, i).
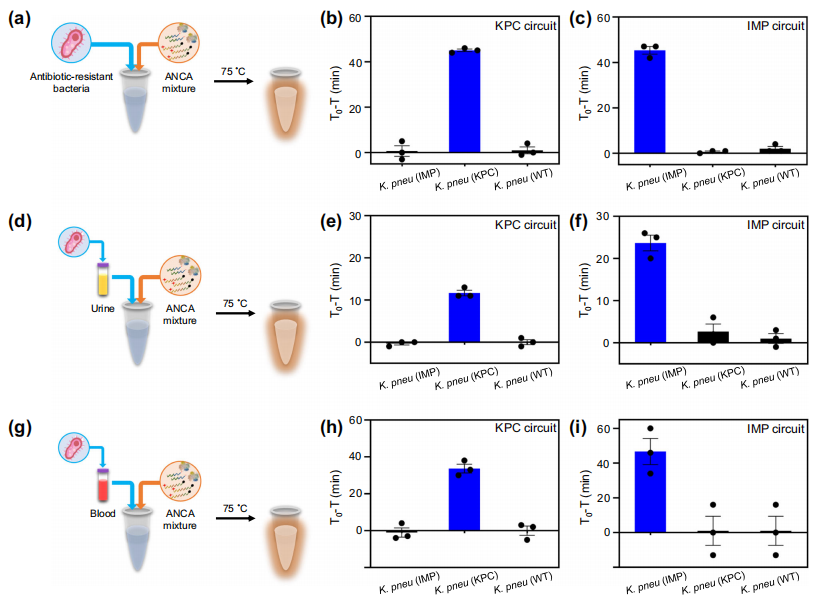
Direct detection of resistant bacteria using the ANCA method. Image source: Nature Communications
Application of ANCA Method for Diagnosing Resistant Bacteria
The authors attempted to diagnose CPKP from clinical samples using the ANCA method. Using cotton swabs, 143 rectal swabs were collected from patients (63 KPC positive, 80 negative), each placed in transport media. The transport media was directly mixed with ANCA reagents and incubated under fluorescent monitoring. The results showed that the ANCA method could distinguish between 63 KPC positive samples and 79 negative samples (Figure b), with only sample 2 detecting a false positive signal, and there was a strong correlation between the ANCA method and PCR method (Figure c). The KPC diagnostic results from a total of 200 clinical and spiked samples showed clinical sensitivity and specificity of 100% and 98.7%, respectively. ROC analysis yielded an area under the curve (AUC) value of 0.9997 (Figure d). The IMP diagnostic results from 160 clinical and simulated clinical specimens (80 IMP positive, 80 IMP negative) showed clinical sensitivity and specificity of 100% and 100%, with an AUC of 1.0 (Figure e).
The authors attempted to directly detect Klebsiella pneumoniae (KPC) from patient swabs using the ANCA method. The results showed that the diagnostic results from rectal swabs (sensitivity = 100%, specificity = 100%) were more accurate than those from the transport media in which the swabs were immersed (sensitivity = 98.7%, specificity = 100%).
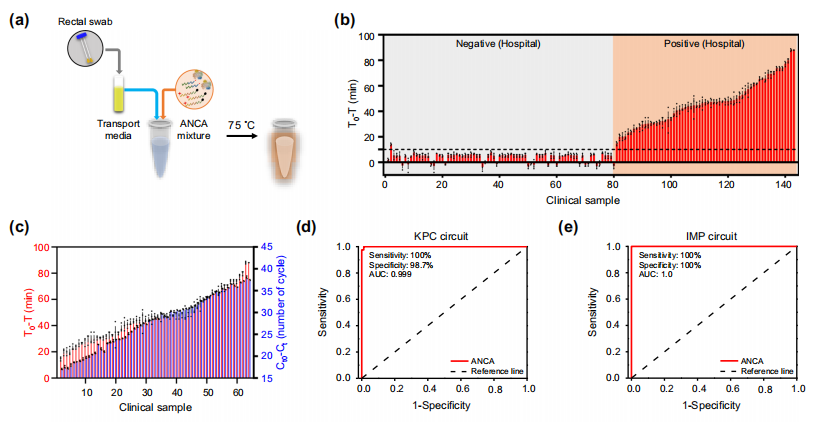
Application of the ANCA method for diagnosing resistant bacteria. Image source: Nature Communications
3D Nanocolumn Swab ANCA Method for Capturing and Detecting Resistant Bacteria
The authors further used the ANCA method to capture and detect antibiotic-resistant bacteria from contaminated surfaces (Figure a). They employed a 3D nanocolumn array swab to attempt to capture bacteria from surfaces such as pig skin, tables, gloves, scissors, knobs, and tweezers. The swabs capturing the bacteria were directly placed into tubes for the ANCA reaction. The complex 3D nanocolumn array structure promotes bacterial adhesion through nanoscopic topological interactions, allowing for irreversible bacterial capture, thus preventing secondary infections. The results of using the ANCA method to detect KPC and IMP for each target showed specific fluorescent signals (Figures c and d).
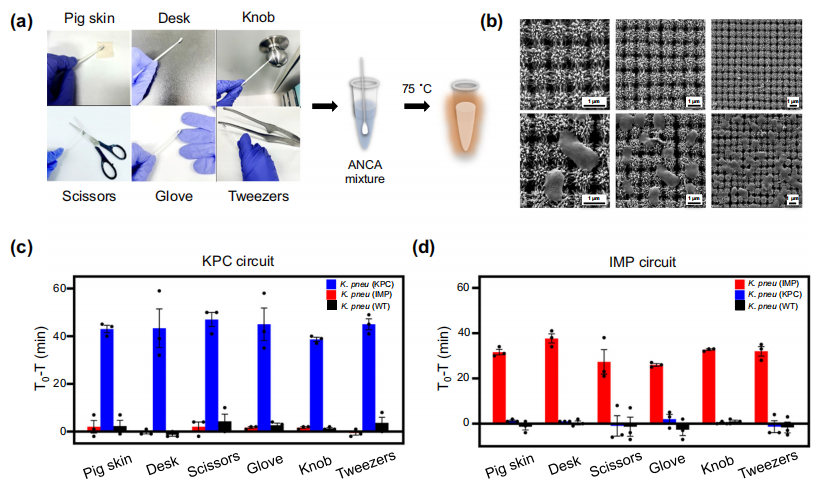
Using ANCA to capture and detect antibiotic-resistant bacteria with a 3D nanocolumn swab method.
Image source: Nature Communications
Conclusion and Discussion
The authors report a molecular diagnostic strategy (ANCA) that achieves one-step, amplification-free, isothermal DNA detection through cross-catalytic cleavage by Ago proteins, successfully detecting carbapenemase-producing Klebsiella pneumoniae (CPKP) without the need for DNA extraction and amplification steps. Furthermore, the authors demonstrated the method’s application in detecting carbapenem-resistant bacteria in human urine and blood samples, as well as directly identifying CPKP from surfaces using a 3D nanocolumn swab. The ANCA method was applied to detect CPKP in rectal swab specimens from infected patients, achieving sensitivity and specificity of 100% and 100%, respectively. This method enables simple, rapid, and accurate diagnosis of CPKP, aiding in the prevention of nosocomial infections.
The ANCA method can directly detect CPKP from rectal swabs collected from patients, and the diagnostic results obtained from rectal swabs prove to be more accurate than those obtained from the transport media in which the swabs were immersed. By using a 3D nanocolumn array swab to collect clinical samples and directly performing ANCA, accurate diagnosis of CPKP can be predicted without the risk of secondary infections, as this method can avoid potential sample contamination and limit bacterial transmission. The next step will carefully evaluate the effectiveness of combining the ANCA method with the 3D nanocolumn array swab. This will involve close cooperation with physicians to enhance the implementation of this method in real-world clinical settings.
Future research avenues may explore improving the cleavage efficiency of Ago proteins/guide DNA. Compared to other techniques, the relatively longer reaction time is a drawback of ANCA. In the absence of guide DNA, Ago proteins often form Apo, leading to non-specific cleavage reactions of dsDNA. To suppress this situation, the concentration of Ago proteins should not be too high. However, the higher the concentration of Ago proteins, the more favorable the reaction kinetics. Thus, the authors chose a moderate concentration of 200 nM Ago proteins to address both issues, hoping this problem will be resolved soon. This expectation is based on the latest advancements in protein engineering, which have led to the emergence of Apo-free forms of Ago proteins. Another drawback is that the data provided in the current NEB DNA design guidelines are highly variable, leading to differences in sensitivity values for each circuit. As more data is collected, it may become possible to detect different target nucleic acids with consistent efficiency.
— Exciting Video Recommendations —
—  —
—

In vitro diagnostic new media & service platform
Satisfy curiosity & solve problems

Long press to joinKnowledge Planet
Clear the fog and light up the future

Long press to identifyCustomer Service WeChat
AddFriends to invite you into the group
Hotline: 4008-0571-82
Email: [email protected]
The views expressed in this reproduced article only represent the original author. If there is any infringement, please contact for deletion!
Click “View” to give me a little orange flower
