Introduction
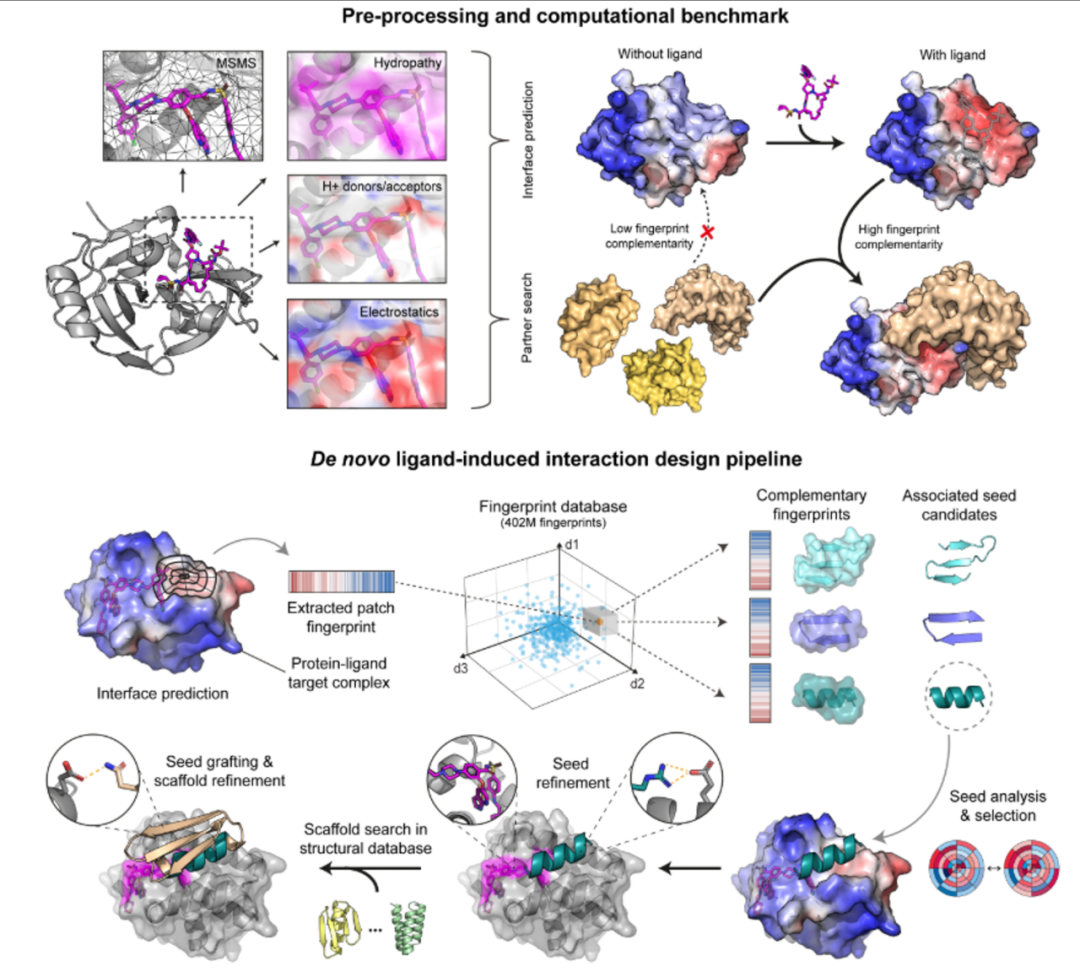
MaSIF-neosurface is a computational tool aimed at learning new surfaces (neosurfaces) of protein-small molecule complexes, designed to identify potential binding regions within the new surface structures formed after protein binding with small molecules. This method proposes a deep learning-based protein design strategy that can be used to develop artificial proteins capable of specifically recognizing these “new surfaces.” The strategy utilizes geometric and chemical fingerprint features of protein surfaces to extract functional feature patterns by training deep models.
MaSIF-neosurface is trained solely on protein surface data, effectively generalizing to new surfaces formed by small molecule binding, demonstrating a rare cross-modal transfer capability in deep learning.
01
Xingchen Create Python Environment and Install Basic Dependencies
conda create -n masif python==3.6 -y conda activate masif
Create a virtual environment named masif, specifying Python version 3.6 (MaSIF depends on this version).
conda install -c open3d-admin open3d=0.5 -y
Install an older version of Open3D (for 3D point cloud processing).
conda install -c conda-forge rdkit openbabel prody biopython dask matplotlib tensorflow=1.9 -y
Install a series of tools for biological structure processing and visualization:
-
rdkit, openbabel: Chemical structure processing
-
prody, biopython: Protein structure analysis
-
dask: Parallel data processing
-
matplotlib: Plotting
-
tensorflow=1.9: MaSIF depends on TensorFlow 1.x version
pip install StrBioInfo pip install nose pip install pdb2pqr==3.5.2
-
StrBioInfo: Structural bioinformatics tool
-
nose: Testing framework
-
pdb2pqr: Used to generate charged structure files from PDB (subsequently used for potential calculations)
02
Xingchen Install PyMesh
git clone https://github.com/PyMesh/PyMesh.git … python build.py all cmake .. make ./setup.py build ./setup.py install python -c “import pymesh; pymesh.test()”
PyMesh is a high-performance mesh manipulation library used for 3D meshing of protein surfaces; MaSIF uses it to generate .ply surface data.
03
Xingchen Install Reduce
git clone https://github.com/rlabduke/reduce.git … make sudo make install
Reduce is a standard tool for adding hydrogen atoms to protein structures (MaSIF requires precise hydrogen locations to construct surface charges).
04
Xingchen Install MSMS (Protein Surface Generation)
mkdir -p msms …https://ccsb.scripps.edu/msms/downloads/
MSMS is a protein surface generation program; MaSIF uses MSMS to construct the “solvent-accessible surface” from PDB.
Note: Manual download of msms_i86_64Linux2_2.6.1.tar is required for extraction and use.
05
Xingchen Install APBS + PDB2PQR
git clone https://github.com/Electrostatics/apbs-pdb2pqr.git cd apbs-pdb2pqr cmake -DGET_MSMS=ON apbs make sudo make install
-
APBS: Calculates protein surface potential
-
PDB2PQR: Preprocesses structures for potential calculations
-
GET_MSMS=ON: Automatically associates with MSMS
06
Xingchen Other Components
conda install -c schrodinger pymol=2.5.0 -y (install different versions as needed)
Install PyMOL for visualizing .ply surfaces.
After installing the plugin masif_pymol_plugin.py, you can load using loadply file.ply.
07
Xingchen Download Benchmark Dataset (Provided by MaSIF)
wget https://zenodo.org/records/14225758/files/masif-neosurf-benchmark-dataset.zip?download=1 wget https://zenodo.org/records/14225758/files/pdbbind_decoys.zip?download=1
Includes training sets, evaluation structures, docking decoy data, etc.
08
Xingchen Environment Variable Configuration (Reference)
export MSMS_BIN=/usr/local/bin/msms export APBS_BIN=/usr/local/bin/apbs export PDB2PQR_BIN=/home/install/miniforge3/envs/masif/bin/pdb2pqr30 export MULTIVALUE_BIN=/usr/local/share/apbs/tools/bin/multivalue export REDUCE_HET_DICT=/home/install/masif-dep/reduce/reduce_wwPDB_het_dict.txt
Official tutorial: GitHub – LPDI-EPFL/MASIF: MaSIF – Molecular Surface Interaction Fingerprints. Geometric deep learning for deciphering patterns in molecular surfaces.
@
Your attention, sharing, and feedback are the driving force for us at Xingchen.
May you shine brightly and continuously on the path of scientific drug research.
